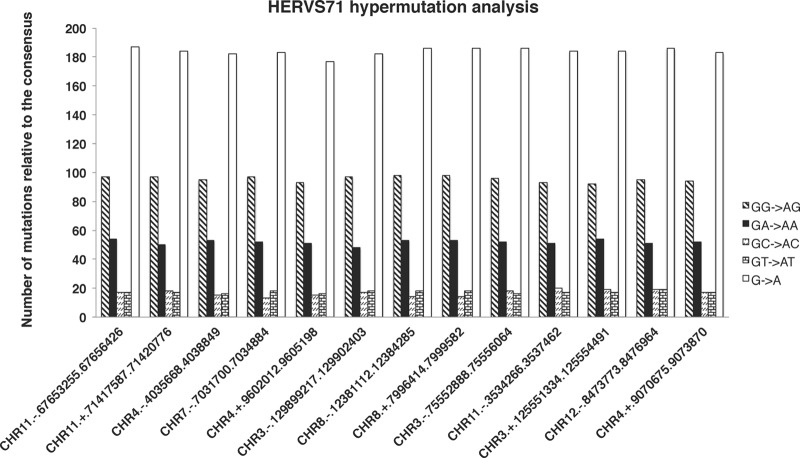
Fig 6.
Analysis of the hypermutation footprint of hA3G and hA3F on 13 HERV-S71 sequences from the ERV-1 family. Sequences were aligned using MAFT, and a consensus sequence was generated from 25 HERV-S71 sequences that had normal DR values. Thirteen suspected hypermutated HERV-S71 sequences were aligned with the consensus sequence, and their hypermutation status was investigated using the Hypermut2 program from the Los Alamos National Laboratory. The vertical axis shows the number of mutations in each sequence against the consensus sequence. The horizontal axis shows a brief description of each of the HERV-S71 sequences (chromosomal location described by the RepeatMasker tag). Each sequence description has chromosome number, strand, starting position, and end position, each separated by a period. The bars represent total numbers of G-to-A mutations as well as those within different sequence contexts.