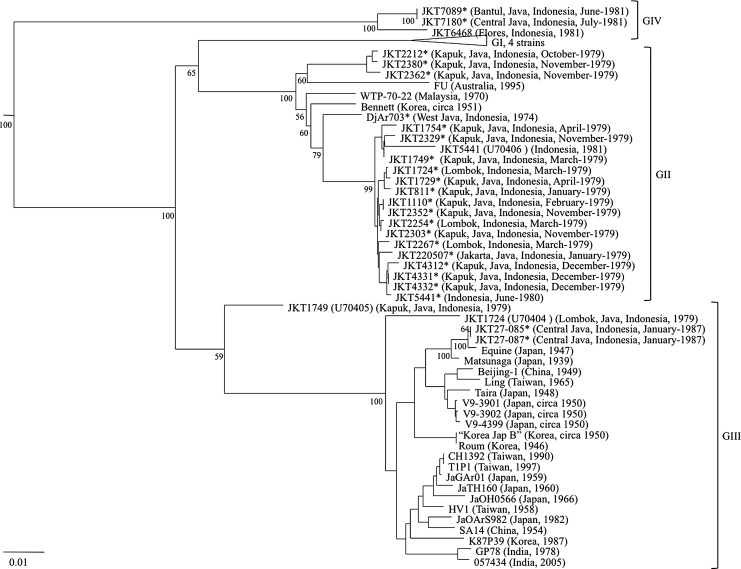
FIG. 2.
Neighbor-joining phylogeny based on nucleotide sequence information derived from the E gene of the Japanese encephalitis virus (JEV) isolates. The tree was rooted using the MVE-1-51 isolate of Murray Valley encephalitis virus, which is a member of the Japanese encephalitis serogroup, but has been removed to allow for better visualization of branch lengths. Horizontal branch lengths are proportional to the genetic distance between strains, and the scale at the lower-left of the tree indicates the number of nucleotide substitutions per site. Genotype (GI–IV) is represented to the right of the tree. Bootstrap percentages based on 1000 replicates are indicated to the lower-left of the genotype-defining nodes within the phylogeny, as well as additional selected nodes. The Indonesian isolates sequenced in this study are indicated by asterisks.