Abstract
During development and cellular differentiation, tissue and cell-specific programs mediate mitochondrial biogenesis in order to meet physiological needs. However, environmental and disease-associated factors can perturb mitochondrial activities requiring cells to adapt to protect mitochondria and maintain cellular homeostasis. Several mitochondria-to-nuclear signaling pathways, or retrograde responses, have been described, but the mechanisms by which mitochondrial stress or dysfunction is sensed to precisely coordinate the appropriate response has only recently begun to be understood. Recent studies of the mitochondrial unfolded protein response (UPRmt) indicate that the cell monitors mitochondrial protein import efficiency as an indicator of mitochondrial function. Here, we review how the cell evaluates mitochondrial function and regulates transcriptional induction of the UPRmt, adapts protein synthesis rates and activates mitochondrial autophagy to promote mitochondrial function and cell survival during stress.
Keywords: Mitochondrial Stress, Transcriptional Responses, Autophagy
Mitochondrial Evolution and Communication with the Nucleus
The endosymbiotic origin of mitochondria that occurred over a billion years ago via the engulfment of a proteobacteria by a eukaryotic precursor cell was instrumental in the evolution of multicellular organisms. Eukaryotic cells gained many advantageous traits as a result including a unique signaling platform to mediate apoptosis and the response to viral infection [1, 2]. But the most important advantage resulting from endosymbiosis is the ability to efficiently generate adenosine 5′ triphosphate (ATP) via aerobic respiration [3]. In addition to the numerous advantages, the presence of the new organelles also presented a number of new challenges including maintenance, replication and biogenesis of the topologically complex compartment.
Mitochondria are double membrane organelles found in nearly all eukaryotic cells. Depending on physiologic need, cells harbor from 10-1000 mitochondria. The mitochondrial proteome is composed of ~1100 proteins which are encoded by genes located in both the nuclear and mitochondrial genomes (mtDNA) [4]. Each mitochondrion contains 3-10 mtDNAs, which encodes 13 components of the electron transport chain (ETC) and the tRNAs and rRNAs required for their synthesis. Thus, the majority of ETC components and mitochondrial proteome is encoded by nuclear genes, translated in the cytosol and imported into each organelle [5].
Because ~99% of mitochondrial proteins are encoded by the nucleus and mitochondrial activities impact many essential cellular processes ranging from metabolism to DNA replication [6], signaling pathways have evolved to communicate between the two organelles to mediate diverse transcriptional responses. Mitochondrial-to-nuclear communication, or retrograde signaling, has been established via the pioneering work from the Butow and Avadhani laboratories [7].
The RTG signaling pathway in Saccharomyces cerevisiae
The RTG response in yeast is a well-characterized signaling pathway activated by ETC deficiency resulting in the transcriptional up-regulation of genes that coordinate metabolic changes to maintain glutamate supplies and resupply mitochondria with oxaloacetate and acetyl-CoA [7]. The physiological importance of the RTG pathway in metabolic remodeling is clear, as ETC deficient cells lacking the RTG genes become glutamate auxotrophs [8, 9].
Three proteins are essential regulators of the RTG response. Rtg1p and Rtg3p are transcription factors that form a complex and upon activation bind to the R box sequence in target gene promoters to stimulate transcription [10]. When off, Rtg1p and Rtg3p are localized in the cytosol where Rtg3p is heavily phosphorylated. However, during ETC dysfunction, Rtg3p becomes less phosphorylated and both Rtg3p and Rtg1p traffic to the nucleus where they are transcriptionally active [11]. Rtg1p and Rtg3p translocation requires the upstream component Rtg2p, which is thought to act as sensor of mitochondrial dysfunction [11, 12]. More recently, it has been suggested that the RTG pathway is triggered by loss of mitochondrial inner membrane potential [13], but how this stimulates Rtg2 or the downstream transcription factors is unclear. There are many negative regulators of the RTG response that potentially integrate RTG signaling with other metabolic activities such as the TOR (target of rapamycin) pathway, but these results are more thoroughly elaborated by others [7].
Mammalian Retrograde Signaling
Proteins homologous to the RTG proteins are not obvious in metazoans but conceptually similar pathways have been demonstrated by a variety of studies indicating alterations in nuclear gene expression during mitochondrial dysfunction [14- 18]. The pioneering studies by Avadhani’s group utilized mtDNA depletion or mutations in nuclear encoded respiratory chain mutants to elucidate a number of retrograde signaling pathways, which are briefly discussed below and in greater detail elsewhere [7, 19].
In this case, retrograde signaling is largely thought to occur in response to cytosolic calcium accumulation that occurs at least partially due to the inability of dysfunctional mitochondria to take up cytosolic calcium [20, 21]. Accumulating cytosolic calcium leads to activation of calcineurin and subsequently the transcription factors NF-κB, CHOP and ERK1, among others [7]. During mitochondrial stress, NF-κB activation occurs independent of cytokines and chemokines and the traditional NF-κB repressor IκBα. Rather, mitochondrial stress causes calcineurin-dependent dephosphorylation of the NF-κB inhibitor IκBβ, allowing NF-κB to traffic from the cytosol to the nucleus [20]. The importance of this signaling pathway has primarily been demonstrated by the gain-of-function it provides cancer cells, which includes increased invasiveness and tumor growth [20, 22].
The Mitochondrial Unfolded Protein Response (UPRmt)
The presence of a separate retrograde response, termed a mitochondrial stress or mitochondrial unfolded protein response (UPRmt) was suggested by the induction of nuclear-encoded mitochondrial chaperones and proteases during mitochondrial stress caused by the accumulation of unfolded or misfolded proteins within the mitochondrial matrix [23, 24]. A similar response was later documented in C. elegans, which was also activated by a number of conditions that cause mitochondrial stress including depletion of the mitochondrial genome [25], ETC genes [26], depletion of mitochondrial proteases [27] and reagents that generate high levels of reactive oxygen species (ROS) [25]. The physiological importance of the UPRmt is demonstrated, as worms lacking UPRmt signaling components have impaired development and survival during conditions that perturb mitochondrial function [27, 28].
Recent microarray experiments from our laboratory have demonstrated a much broader transcriptional scope of the UPRmt in addition to the known nuclear encoded mitochondrial molecular chaperones and proteases that promote protein folding or clearance of defective proteins within stressed mitochondria [29]. Additionally, the UPRmt includes ROS scavenging machinery, mitochondrial fission genes, genes involved in ubiquinone biosynthesis and iron-sulfur cluster biogenesis as well as glycolysis [27]. These results suggest that in addition to promoting mitochondrial protein homeostasis, the UPRmt also adapts metabolism to allow the cell to better withstand mitochondrial stress (see below).
Intra-cellular Evaluation of Mitochondrial Function and UPRmt Activation
As mitochondrial dysfunction is pleiotropic and separating mitochondrial dysfunction from mitochondrial unfolded protein accumulation is challenging, pin-pointing the mean(s) used by the cell to evaluate mitochondrial function and coordinate UPRmt induction has been difficult. Because, dysfunction results in activation of a downstream transcriptional response, the monitoring mechanism must be able to communicate activities of the intra-mitochondrial compartments with the cytosol and ultimately the nucleus. Recent studies from our laboratory indicate that the cell monitors mitochondrial protein import efficiency to determine mitochondrial function and appropriately activate the UPRmt [27].
Mitochondrial Protein Import Efficiency
Mitochondrial protein import is a well-characterized process dependent on a variety of mitochondrial activities [30] making it a potentially useful pathway to evaluate mitochondrial function. Once synthesized in the cytosol, nuclear-encoded proteins are targeted to mitochondria by signal sequences as loosely folded precursors. Import into mitochondria requires the TOM (Translocase of the Outer Membrane) and TIM (Translocase of the Inner Membrane) complexes [30, 31]. Substrates destined for the mitochondrial matrix typically have an amino-terminal mitochondrial targeting sequence (MTS), which forms an amphipathic helix. The MTS first interacts with the TOM channel on the cytosolic surface of mitochondria and subsequently with the TIM23 complex to cross the inner membrane and enter the matrix. Once exposed to the matrix, the MTS is cleaved allowing the protein to fold or assemble with the assistance of mitochondria-specific molecular chaperones including Hsp60 and mtHsp70 [30].
Traversing the inner membrane requires the ETC-generated membrane potential, which facilitates the import of the positively charged MTS via the electrophoretic effect during translocation [32]. Additionally, import requires mtHsp70, which functions both as a motor in the PAM (presequence translocase-associated motor) complex to transport precursor proteins across the inner membrane [33] and to promote protein folding in the matrix. A number of general perturbations to mitochondria are known to slow or impair import including the ATP synthase inhibitor oligomycin [34], the NADH-ubiquinone oxidoreductase inhibitor rotenone [35] and paraquat, which causes excess ROS generation [36] in addition to inhibition of mitochondrial chaperones [37]. It is the requirement for numerous mitochondrial activities that makes import efficiency a useful readout for mitochondrial function.
Coordination of mitochondrial stress with UPRmt induction
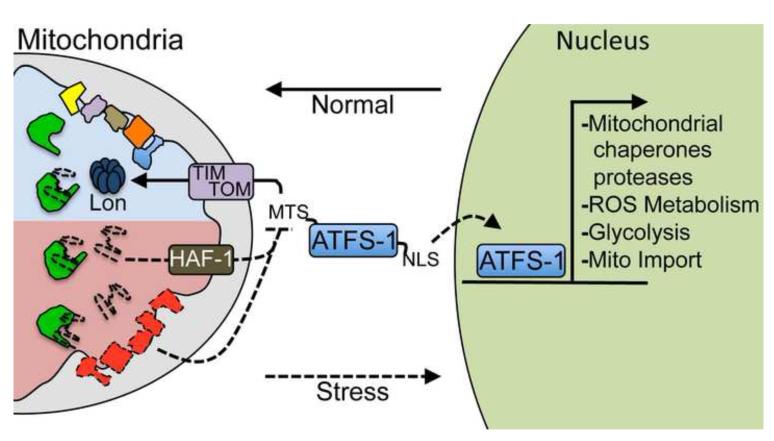
Recent work from our laboratory has indicated the requirement for Activating Transcription Factor associated with Stress-1 (ATFS-1) in UPRmt induction [27, 28]. In addition to a nuclear localization sequence (NLS) in the leucine zipper domain, ATFS-1 also has an amino-terminal MTS. The unique ability to localize to both compartments potentially enables ATFS-1 to function as a sensor of mitochondrial import efficiency. In the absence of mitochondrial stress or UPRmt induction, ATFS-1 is imported into mitochondria and degraded by the Lon protease (Figure 1), presumably as a negative regulatory mechanism [27].
Figure 1. The mitochondrial unfolded protein response.
An illustration of the signaling mechanism that regulates the induction of the mitochondrial unfolded protein response (UPRmt) as elucidated in C. elegans. The UPRmt is activated during mitochondrial dysfunction or stress resulting in the transcriptional up-regulation of protective genes including mitochondrial chaperones and proteases, those involved in reactive oxygen species (ROS) detoxification, the glycolysis pathway and the mitochondrial protein import machinery. The cell determines mitochondrial function and when it is appropriate to induce the UPRmt by monitoring the mitochondrial protein import efficiency of the transcription factor ATFS-1. In the absence of mitochondrial stress, ATFS-1 is translated and efficiently imported into mitochondria via a mitochondrial targeting sequence (MTS), where ATFS-1 is degraded by the Lon protease. However, during mitochondrial dysfunction, general protein import efficiency is reduced allowing a percentage of ATFS-1 to accumulate in the cytosol. Because ATFS-1 also has a nuclear localization sequence (NLS), it then traffics to the nucleus where it induces the UPRmt. Mitochondrial import efficiency can be impaired or reduced by a number of conditions including mitochondrial chaperone depletion or respiratory chain dysfunction. Additionally, import can be slowed by peptide efflux via the ABC transporter HAF-1, which occurs when the mitochondrial chaperone (green) capacity is exceeded by unfolded proteins (dashed lines); all of which result in UPRmt induction to maintain organelle homeostasis.
During mitochondrial dysfunction when UPRmt signaling occurs, a small percentage of ATFS-1 fails to be imported into mitochondria and accumulates in the cytosol suggesting that mitochondrial import is impaired [27, 28]. Additional mitochondrial proteins were also found in the cytosol suggesting a general impairment of mitochondrial import during stress, but because ATFS-1 also has a NLS it is able to traffic to the nucleus to induce the UPRmt (Figure 1). Consistent with the cell monitoring mitochondrial import efficiency via ATFS-1, any condition that perturbs import including inhibition of the TOM and TIM complexes, ETC perturbation, paraquat treatment and mitochondrial chaperone inhibition caused ATFS-1-dependent UPRmt induction. Furthermore, preventing mitochondrial import of ATFS-1 by simply deleting the MTS was sufficient to cause ATFS-1 nuclear accumulation and UPRmt induction even in the absence of mitochondrial dysfunction [27].
Dual localization of a protein to the cytosol and mitochondrial matrix has been documented for several proteins including the enzyme fumarase [38]. Interestingly, the MTS from both the mitochondrial and cytosol-localized forms of fumarase is cleaved in the matrix. If the C-terminal domain remains unfolded during mitochondrial import the MTS is cleaved and the entire protein enters the matrix where it folds and functions in the tricarboxylic acid cycle. However, if the C-terminal domain folds prior to being completely imported, the MTS is cleaved, but the remainder of the polypeptide diffuses back into the cytosol [39]. In this respect, fumarase localization depends on import efficiency although the process is thought to be stochastic as fumarase accumulates in both compartments under normal physiologic conditions.
Additionally, it is not unprecedented that the regulation of a stress-associated transcription factor be tightly linked to its degradation. Both p53 and Hif1 are constitutively translated, recognized by a regulatory ubiquitin ligase, and ultimately degraded by the cytosolic or nuclear pool of proteasomes [40, 41]. Only in the presence of stress such as DNA damage or hypoxia does each transcription factor avoid degradation and activate their respective transcriptional responses. However, coupling of degradation with mitochondrial import as a means to control transcription factor activity appears to be a novel means of mitochondria-to-nucleus communication dependent on ATFS-1 having dual organelle localization signals.
Once in the nucleus, ATFS-1 activates a relatively broad transcriptional response to alleviate the effects of mitochondrial stress. The transcriptional response mediated by ATFS-1 during mitochondrial stress is consistent with what might be expected of a mitochondrial stress response or a UPRmt. As expected, ATFS-1 induced the expression of the known mitochondrial chaperone genes as well as several mitochondrial proteases that re-establish protein homeostasis as well as restore import efficiency. Additionally, the UPRmt induced a number of ROS-scavenging components that are localized to both the mitochondria and the cytosol consistent with increased ROS production during mitochondrial stress. Interestingly, a number of genes in the glycolysis pathway were also up-regulated suggesting that ATFS-1 may shift metabolism from mitochondrial-dependent respiration to glycolysis as an alternate means of ATP production in the presence of mitochondrial dysfunction.
Lastly, ATFS-1 mediates the up-regulation of two core components of the TIM23 complex; tim-17 and tim-23, which are required for the import of N-terminal MTS containing proteins to the mitochondrial matrix [30] such as ATFS-1 and the mitochondrial targeted molecular chaperones that ATFS-1 transcriptionally up-regulates. The increase in mitochondrial protein import machinery highlights the link between ATFS-1 and mitochondrial import efficiency and suggests a possible means to increase import of protective proteins such as chaperones as well as to down-regulate the UPRmt once stress has been alleviated by improving mitochondrial import and thus preventing ATFS-1 from reaching the nucleus.
Regulation of import and the UPRmt during stress by a mitochondrial peptide exporter
Defects that directly impair mitochondrial protein import such as respiratory chain and mitochondrial chaperone inhibition are severe resulting in developmental impairment and UPRmt induction. In this scenario, the cell is responding to damage that may be unrecoverable and the UPRmt is unable to provide much protection. Is there a mechanism to sense relatively minor mitochondrial damage and activate the UPRmt early on during mitochondrial stress to prevent the further accumulation of damage? In addition to ATFS-1, we identified HAF-1, a mitochondrial inner membrane-localized ABC peptide transporter as being a modulator of UPRmt induction [27, 28]. HAF-1 is only required for UPRmt induction during the initial stages of what will eventually become severe stress such as treatment with ethidium bromide which impairs mtDNA expression or during mild mitochondrial stress that does not cause developmental arrest such as hypomorphic mutations in ETC components.
Interestingly, our data suggest that HAF-1 serves to slow general mitochondrial protein import during mild mitochondrial stress. During mild stress in haf-1-deletion worms, ATFS-1 as well as other substrates fail to accumulate in the cytosol or nucleus, rather they continue to accumulate in mitochondria explaining why the UPRmt is impaired in the absence of HAF-1. However, HAF-1 is not required during UPRmt activation caused by direct import impairment or when the MTS is removed from ATFS-1, consistent with the proposed role as a modulator of mitochondrial import that attenuates import and activates the UPRmt to prevent severe damage accumulation. Previous work from our laboratory and others suggests that HAF-1 pumps peptides derived from the degradation of misfolded proteins within the mitochondrial matrix linking its activity to the accumulation of unfolded proteins although it is currently unclear which HAF-1 peptide substrate(s) affect protein import or by what mechanism [28, 42].
We hypothesize that HAF-1-dependent import attenuation evolved independently of the UPRmt for two reasons. First, a homologous mitochondrial peptide transporter exists in S. cerevisiae, Mdl1p [42] without an obvious ATFS-1 ortholog [43]. It should be noted that the role of Mdl1 in the regulation of mitochondrial import has not been examined. Second, slowing import during organelle stress potentially reduces the burden on the mitochondrial protein folding machinery allowing the folding environment to re-establish homeostasis before introducing additional substrates. Analogous cellular strategies are employed during stress in the endoplasmic reticulum to reduce import via translation attenuation (see below) [44].
We have found that HAF-1 functions upstream of ATFS-1 to regulate the UPRmt. As HAF-1′s effect on mitochondrial import is not specific to ATFS-1, it will be interesting to discover if other transcription factors or signaling pathways are regulated in a similar manner. The signaling mechanism regulating the UPRmt has largely been elucidated in C. elegans, although similar transcriptional responses have been observed during mitochondrial stress in mammalian systems [24, 45, 46]. Going forward, it will be important to determine if a similar mechanism regulates the UPRmt in mammals and its role during aging and diseases associated with mitochondrial dysfunction (Boxes 1 and 2).
Box 1. Mitochondrial Stress Responses in Aging.
Studies in multiple organisms indicate that mitochondria are intimately associated with the aging process. A decline in mitochondrial function as cells age has been well documented and manipulations that increase mitochondrial dysfunction such as increasing mutation accumulation in the mitochondrial genome can cause phenotypes consistent with accelerated aging [75, 76]. Interestingly, by increasing mitochondrial biogenesis, via overexpression of the transcriptional regulator PGC-1α, several age-associated phenotypes can be delayed [77], consistent with mitochondrial decline contributing to aging and suggesting that strategies to maintain mitochondrial function may be of therapeutic value.
These results also suggest that a system or systems may exist to promote mitochondrial biogenesis or function in aging cells to prevent or slow the decline in mitochondrial function over time thus counteracting the normal aging process. However, the GCN2-mediated pathway or the UPRmt do not appear to be employed to mitigate damage associated with normal aging at least in yeast and worms as impairment of GCN2 signaling or UPRmt activation has little or no effect on the lifespan of otherwise wild-type strains [26, 55, 74]. However, emerging results indicate that if stimulated, the GCN2 pathway and the UPRmt can contribute to lifespan extension potentially delaying or slowing the aging process. Somewhat paradoxically, a number of mutations that cause mitochondrial dysfunction including defects in respiratory chain subunits or the m-AAA protease extend lifespan in organisms such as yeast and worms [26, 49, 78]. These same conditions activate the GCN2 translation attenuation pathway as well the UPRmt and require both mitochondrial stress response pathways for the observed extension of lifespan. However, because many signaling pathways likely emanate from mitochondria it will be interesting to determine if the UPRmt is also sufficient to prolong lifespan. Impressively, work in yeast indicates that increased Gcn4p (an output of GCN2-mediated eIF2α phosphorylation (Figure 2)) is sufficient to extend lifespan [74]. It will be of interest in the future to determine if the lifespan extension or delayed aging is due to direct effects on mitochondria such as increased biogenesis or reduced degeneration or if the stress response pathways also activate compensatory mechanism that reduce the necessity for mitochondrial activities during aging such as through altered metabolism [79].
Box 2. Parkinson’s Disease and Mitochondrial Stress Signaling.
Parkinson’s disease (PD) is a degenerative disorder resulting largely from the death of dopaminergic neurons in the substantia nigra region of the brain. The etiology of PD is complex and likely influenced by many environmental and genetic factors. However, years of research strongly suggests that mitochondrial dysfunction plays an important role in PD progression suggesting a potentially protective role for the mitochondrial stress response pathways described. A variety of ETC inhibitors such as rotenone [80] and paraquat have been shown to cause PD or induce PD-like symptoms. Furthermore, several loss-of-function mutations that affect mitochondrial biology are known to cause juvenile onset Parkinson’s disease including mutations in the genes encoding PINK1 and Parkin [81, 82]. PINK1 and Parkin are involved in numerous processes including the mitophagy pathway, which specifically recognizes and targets severely dysfunctional mitochondria for degradation [66]. A role for PINK1-dependent mitophagy is expected in dopaminergic neurons because PINK1 loss-of-function results in dopaminergic cell death but mitophagy has been difficult to document in vivo [83]. A role for the UPRmt or GCN2 has not been examined in dopaminergic neurons but is possible because the ETC poisons that elicit PD-like phenotypes are strong UPRmt inducers and the requirement for PINK1 suggests a defect in mitochondrial protein import which is also known to activate the UPRmt [27]. Interestingly, a reduction in protein synthesis rescues many of the defects observed in flies lacking PINK1 suggesting a protective role for translation attenuation during conditions associated with PD [84]. Future studies will be important to determine the role, if any, of the ATFS-1 and GCN2-dependent mitochondrial stress response pathways in the protection of dopaminergic neurons during conditions that lead to PD.
Translation Attenuation During Mitochondrial Dysfunction
Several recent reports have suggested that reduced cytosolic protein synthesis is protective during mitochondrial dysfunction. For example, mutations in the adenine nucleotide translocase cause age associated mitochondrial degeneration, which is exacerbated by impairment of mitochondrial protein quality control machinery. Interestingly, reagents or mutations that slowed cytosolic translation or protein synthesis were protective significantly delaying the onset of mitochondrial dysfunction most likely by protecting the mitochondrial protein-folding environment [47].
More recently, mitochondrial dysfunction was shown to directly cause the attenuation of cytosolic protein synthesis as discovered in mammalian cells treated with the ATP synthase inhibitor oligomycin and in yeast by comparing the stress resistance profiles of yeast strains lacking a mitochondrial quality control protease to those lacking subunits of the large cytosolic ribosome subunit [48, 49]. Interestingly, strains lacking ribosomal components or the mitochondrial protease were resistant to similar stresses including mitochondrial stress caused by paraquat and endoplasmic reticulum (ER) stress caused by tunicamycin. All of the ribosomal mutants had reduced protein synthesis rates as did the strain lacking the mitochondrial protease suggesting a protective signaling mechanism is in place to reduce protein synthesis during mitochondrial stress. It has been known for years that reduced protein synthesis protects ER function during stress by reducing the burden of incoming substrates on the protein-folding environment [50]. ER stress is linked to reduced translation rates via the ER membrane-localized protein kinase PERK, which is activated during ER stress and phosphorylates the translation initiation factor eIF2α, thus inhibiting protein synthesis [51]. In sum, these data suggest that increased eIF2α phosphorylation is protective during mitochondrial dysfunction (Figure 2), similarly to that observed during ER dysfunction.
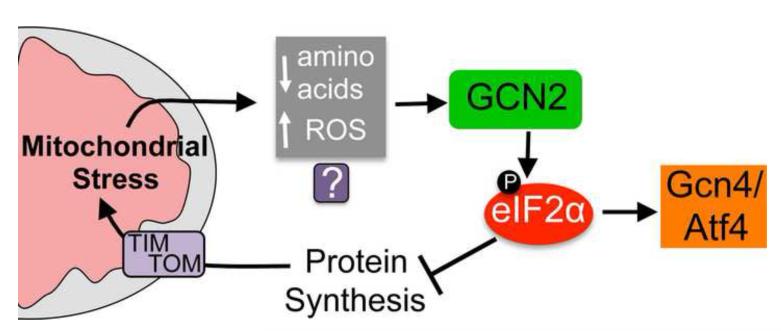
Figure 2. The mitochondrial stress-stimulated translation attenuation pathway.
A model demonstrating how protein synthesis rates are mediated during mitochondrial stress. Conditions including inhibition of a mitochondrial protease required for respiratory chain quality control and mitochondrial ribosome biogenesis [49, 55], respiratory chain or ATP synthase impairment [48] as well as paraquat treatment [55] result in phosphorylation of the translation initiation factor eIF2α by the kinase GCN2. The subsequent decrease in protein synthesis potentially reduces the burden on the protein folding and respiratory chain complex assembly machinery, thus protecting the protein-folding environment. In addition to reducing global protein synthesis, phosphorylated eIF2α promotes the translation and thus activation of the transcription factor Gcn4p in yeast and Atf4 in mammals, both of which are also protective during mitochondrial stress [48, 49, 74]. It is currently unclear how GCN2 becomes activated or senses mitochondrial dysfunction. But, because GCN2 is known to respond to amino acid deprivation and reactive oxygen species (ROS) and both occur during mitochondrial dysfunction multiple possibilities exist.
In addition to global translation attenuation, eIF2α phosphorylation also leads to increased translation of mRNAs with small upstream open reading frames including the transcription factor Gcn4p in yeast and Atf4 in mammals [52, 53]. Oligomycin treatment caused increased Atf4 expression in mammalian cells [48], and Gcn4p was required for the resistance to paraquat in yeast cells [49] indicating that eIF2α phosphorylation is protective for two reasons. Reduced protein synthesis provides relief to the stressed mitochondrial protein-folding environment and because of preferential transcription factor translation and induction of a protective transcriptional program [54].
Our laboratory and others recently discovered that the increased eIF2α phosphorylation that occurs during mitochondrial dysfunction requires the kinase GCN2 (General Control Non-derepressible 2) [48, 55] or the kinase PKR (Protein Kinase, RNA activated) [46, 56]. Consistent with eIF2α phosphorylation and reduced protein synthesis being protective during mitochondrial stress, the development of mutant worms with perturbed mitochondrial function is impaired in the absence of GCN2, but not the ER stress responsive kinase PERK. Furthermore, worms lacking both GCN2 and ATFS-1 were much more sensitive to mitochondrial stress than worms lacking either gene suggesting that GCN2-mediated translation attenuation and ATFS-1- mediated UPRmt induction act in parallel to protect against mitochondrial dysfunction [55]. While ATFS-1 is activated by mitochondrial protein import deficiency, it is currently unclear how GCN2 becomes activated or senses mitochondrial dysfunction. But GCN-2 is known to be activated by amino acid starvation by binding to uncharged tRNAs [57] or by ROS, through an unknown mechanism [58], both of which can occur during mitochondrial dysfunction (Figure 2).
PINK1-mediated Mitochondrial Autophagy
In addition to UPRmt activation and reduced protein synthesis, the PINK1-mediated mitochondrial autophagy (mitophagy) pathway can also be activated during mitochondrial dysfunction. Mitophagy is a means by which cells specifically target dysfunctional mitochondria for lysosome-dependent degradation [59, 60]. Roles for PINK1 and Parkin in addition to mitophagy have been described elsewhere [61, 62] but here we focus on their role in mitophagy to highlight the similarities between the means of regulation of the ATFS-1-mediated UPRmt and PINK1-mediated mitophagy as the mechanism cells utilize to detect and target defective mitochondria are at least conceptually similar to that utilized to mediate the UPRmt. Mitochondrial protein import efficiency of the transcription factor ATFS-1 is the key determinant in the regulation of the UPRmt, which is quite similar to a mechanism cells utilize the kinase PINK1 to recognize and target severely defective mitochondria for degradation via mitophagy as reviewed in detail by others [63]. Elegant work in mammalian cell culture indicates that PINK1 is imported into healthy mitochondria across the outer and inner membrane, processed and degraded [64-67]. However, if the mitochondrial inner membrane potential is dissipated by uncouplers or by stressors such as paraquat or mtDNA mutations [68], PINK1 import is impaired allowing it to integrate and accumulate in the outer mitochondrial membrane [66]. Through this mechanism, PINK1 only accumulates on defective mitochondria where it interacts with the ubiquitin ligase Parkin and leads to the subsequent recruitment of the autophagy machinery and degradation of the defective organelle [62, 66]. While flies and mammals lacking PINK1 and Parkin display mitochondrial dysfunction [69-71], consistent with a role of both in mitochondrial biology, evidence of mitophagy in vivo similar to that observed in cell culture has thus far not been observed [72]. It will be interesting to determine if a similar mitophagy mechanism occurs in vivo and how the functions of PINK1 and Parkin integrate with the other mitochondrial stress responses described here.
Integration of the Mitochondrial Stress Pathways and Future Perspectives
We have described several mitochondrial stress response pathways that promote global mitochondrial function and cell survival during mitochondrial stress. Each pathway has been studied largely independently in specific model systems that allowed for efficient elucidation of individual signaling pathway components. Because all of the pathways are apparently activated by similar conditions, it will be interesting to determine the precise order of pathway activation during stress and if there is negative or positive cross-talk between each pathway to coordinate the cellular response to maintain mitochondrial homeostasis.
It has been shown in C. elegans that ATFS-1 and GCN2 function in parallel and complementary pathways during mitochondrial stress as the absence of GCN2 causes further activation of the ATFS-1 pathway (and vice versa), but the mitophagy has not been examined in this context [73]. One might expect the ATFS-1 and GCN-2-mediated pathways to be activated prior to the mitophagy pathway or during less severe forms of stress to rectify mitochondrial dysfunction prior to the elimination entire organelles via mitophagy, although this has yet to be determined. Alternatively, there may be cell or tissue specific aspects to each signaling pathway that have yet to be identified.
The UPRmt and PINK1-dependent mitophagy pathways appear to both be regulated at the level of mitochondrial import efficiency [25, 66]. Because both are activated by similar stresses, how does the UPRmt and PINK1 coordinate to most efficiently promote global mitochondrial function while removing those organelles that are damaged beyond repair? The UPRmt can be activated when there is a modest reduction import efficiency when only a small percentage of ATFS-1 fails to be imported into mitochondria [27] while PINK1 accumulation on the mitochondrial outer membrane requires conditions that strongly impair import such as inner membrane uncoupling [64-67]. In this case, the cell likely biases towards UPRmt activation when mitochondrial dysfunction is modest as PINK1 is still imported and degraded. But when organellar dysfunction becomes more severe and PINK1 accumulates on the outer membrane of the most damaged organelles both pathways are activated with only the most damaged mitochondria targeted for degradation.
While still at the early stages, a major goal going forward will be to determine if any of these pathways can be manipulated to mitigate the defects caused by mitochondrial dysfunction during aging or disease and developed into therapeutic targets (Boxes 1 and 2). Recent results in model organisms and tissue culture are encouraging in this regard. For example, in a tissue culture model, hyper-activation of the mitophagy pathway was able to reduce the presence of deleterious mutant mitochondrial genomes, in effect preserving the most-healthy organelles [68]. Furthermore, both the UPRmt and the GCN2-dependent translation attenuation pathways have been shown to be involved in the lifespan extension caused by specific forms of mitochondrial dysfunction in both yeast and worms suggesting that manipulation of these pathways may provide physiological benefits [26, 49, 55] (Box 1). In the future it will be important to determine if these pathways are protective against diseases associated with mitochondrial dysfunction as was recently reported in a model of inflammatory bowel [46, 56] and develop strategies to engage or further activate these pathways independent of mitochondrial dysfunction.
Acknowledgements
C.J.F. is supported by an NIH training grant from the NIGMS 5T32GM008539-17. C.M.H. is supported by the Louis V. Gerstner, Jr. Young Investigators Fund, the Alfred W. Bressler Scholar Fund, the Ellison Medical Foundation, and NIH grant from the NIA R01AG040061.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Galluzzi L, et al. Mitochondria: master regulators of danger signalling. Nature reviews. Molecular cell biology. 2012;13:780–788. doi: 10.1038/nrm3479. [DOI] [PubMed] [Google Scholar]
- 2.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467:929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- 4.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt O, et al. Mitochondrial protein import: from proteomics to functional mechanisms. Nature reviews. Molecular cell biology. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 6.Veatch JR, et al. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137:1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 8.Liu Z, Butow RA. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol Cell Biol. 1999;19:6720–6728. doi: 10.1128/mcb.19.10.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- 10.Jia Y, et al. A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol. 1997;17:1110–1117. doi: 10.1128/mcb.17.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekito T, et al. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol Biol Cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z, et al. Retrograde signaling is regulated by the dynamic interaction between Rtg2p and Mks1p. Mol Cell. 2003;12:401–411. doi: 10.1016/s1097-2765(03)00285-5. [DOI] [PubMed] [Google Scholar]
- 13.Miceli MV, et al. Loss of mitochondrial membrane potential triggers the retrograde response extending yeast replicative lifespan. Frontiers in genetics. 2011;2:102. doi: 10.3389/fgene.2011.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crimi M, et al. Skeletal muscle gene expression profiling in mitochondrial disorders. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2005;19:866–868. doi: 10.1096/fj.04-3045fje. [DOI] [PubMed] [Google Scholar]
- 15.Elstner M, Turnbull DM. Transcriptome analysis in mitochondrial disorders. Brain research bulletin. 2012;88:285–293. doi: 10.1016/j.brainresbull.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 16.Marusich MF, et al. Expression of mtDNA and nDNA encoded respiratory chain proteins in chemically and genetically-derived Rho0 human fibroblasts: a comparison of subunit proteins in normal fibroblasts treated with ethidium bromide and fibroblasts from a patient with mtDNA depletion syndrome. Biochimica et biophysica acta. 1997;1362:145–159. doi: 10.1016/s0925-4439(97)00061-6. [DOI] [PubMed] [Google Scholar]
- 17.Tyynismaa H, et al. Mitochondrial myopathy induces a starvation-like response. Hum Mol Genet. 2010;19:3948–3958. doi: 10.1093/hmg/ddq310. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Morais R. Up-regulation of nuclear genes in response to inhibition of mitochondrial DNA expression in chicken cells. Biochimica et biophysica acta. 1997;1352:325–334. doi: 10.1016/s0167-4781(97)00035-3. [DOI] [PubMed] [Google Scholar]
- 19.Ryan MT, Hoogenraad NJ. Mitochondrial-nuclear communications. Annu Rev Biochem. 2007;76:701–722. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- 20.Biswas G, et al. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 1999;18:522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amuthan G, et al. Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. EMBO J. 2001;20:1910–1920. doi: 10.1093/emboj/20.8.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang W, et al. Silencing of IkBbeta mRNA causes disruption of mitochondrial retrograde signaling and suppression of tumor growth in vivo. Carcinogenesis. 2012;33:1762–1768. doi: 10.1093/carcin/bgs190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinus RD, et al. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. European journal of biochemistry / FEBS. 1996;240:98–103. doi: 10.1111/j.1432-1033.1996.0098h.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Q, et al. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoneda T, et al. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. Journal of cell science. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 26.Durieux J, et al. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nargund AM, et al. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haynes CM, et al. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aldridge JE, et al. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PloS one. 2007;2:e874. doi: 10.1371/journal.pone.0000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chacinska A, et al. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill K, et al. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins [see comment] Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- 32.van der Laan M, et al. Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat Cell Biol. 2007;9:1152–1159. doi: 10.1038/ncb1635. [DOI] [PubMed] [Google Scholar]
- 33.Wiedemann N, et al. Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J Cell Biol. 2007;179:1115–1122. doi: 10.1083/jcb.200709087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eilers M, et al. Both ATP and an energized inner membrane are required to import a purified precursor protein into mitochondria. EMBO J. 1987;6:1073–1077. doi: 10.1002/j.1460-2075.1987.tb04860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho LH, et al. Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in Arabidopsis. Plant Physiol. 2008;147:1858–1873. doi: 10.1104/pp.108.121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright G, et al. Oxidative stress inhibits the mitochondrial import of preproteins and leads to their degradation. Experimental cell research. 2001;263:107–117. doi: 10.1006/excr.2000.5096. [DOI] [PubMed] [Google Scholar]
- 37.Kang PJ, et al. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348:137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- 38.Yogev O, et al. Fumarase: a paradigm of dual targeting and dual localized functions. FEBS J. 278:4230–4242. doi: 10.1111/j.1742-4658.2011.08359.x. [DOI] [PubMed] [Google Scholar]
- 39.Sass E, et al. Folding of fumarase during mitochondrial import determines its dual targeting in yeast. J Biol Chem. 2003;278:45109–45116. doi: 10.1074/jbc.M302344200. [DOI] [PubMed] [Google Scholar]
- 40.Haupt Y, et al. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 41.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 42.Young L, et al. Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science. 2001;291:2135–2138. doi: 10.1126/science.1056957. [DOI] [PubMed] [Google Scholar]
- 43.Arnold I, et al. Evidence for a novel mitochondria-to-nucleus signalling pathway in respiring cells lacking i-AAA protease and the ABC-transporter Mdl1. Gene. 2006;367:74–88. doi: 10.1016/j.gene.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 44.Harding HP, et al. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 45.Horibe T, Hoogenraad NJ. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PloS one. 2007;2:e835. doi: 10.1371/journal.pone.0000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rath E, et al. Induction of dsRNA-activated protein kinase links mitochondrial unfolded protein response to the pathogenesis of intestinal inflammation. Gut. 2012;61:1269–1278. doi: 10.1136/gutjnl-2011-300767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, et al. Reduced cytosolic protein synthesis suppresses mitochondrial degeneration. Nat Cell Biol. 2008;10:1090–1097. doi: 10.1038/ncb1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Reyes I, et al. AMPK and GCN2-ATF4 signal the repression of mitochondria in colon cancer cells. The Biochemical journal. 2012;444:249–259. doi: 10.1042/BJ20111829. [DOI] [PubMed] [Google Scholar]
- 49.Delaney JR, et al. Stress profiling of longevity mutants identifies Afg3 as a mitochondrial determinant of cytoplasmic mRNA translation and aging. Aging cell. 2013;12:156–166. doi: 10.1111/acel.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 51.Harding HP, et al. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 52.Dever TE, et al. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- 53.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harding HP, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 55.Baker BM, et al. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS Genet. 2012;8:e1002760. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rath E, Haller D. Mitochondria at the interface between danger signaling and metabolism: role of unfolded protein responses in chronic inflammation. Inflammatory bowel diseases. 2012;18:1364–1377. doi: 10.1002/ibd.21944. [DOI] [PubMed] [Google Scholar]
- 57.Hinnebusch AG. The eIF-2 alpha kinases: regulators of protein synthesis in starvation and stress. Semin Cell Biol. 1994;5:417–426. doi: 10.1006/scel.1994.1049. [DOI] [PubMed] [Google Scholar]
- 58.Mascarenhas C, et al. Gcn4 is required for the response to peroxide stress in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:2995–3007. doi: 10.1091/mbc.E07-11-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim I, et al. Selective degradation of mitochondria by mitophagy. Archives of biochemistry and biophysics. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell death and differentiation. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin SM, Youle RJ. PINK1- and Parkin-mediated mitophagy at a glance. Journal of cell science. 2012;125:795–799. doi: 10.1242/jcs.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 65.Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS biology. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suen DF, et al. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clark IE, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 70.Gispert S, et al. Parkinson phenotype in aged PINK1-deficient mice is accompanied by progressive mitochondrial dysfunction in absence of neurodegeneration. PloS one. 2009;4:e5777. doi: 10.1371/journal.pone.0005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park J, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 72.Sterky FH, et al. Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12937–12942. doi: 10.1073/pnas.1103295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Samann J, et al. Caenorhabditits elegans LRK-1 and PINK-1 act antagonistically in stress response and neurite outgrowth. J Biol Chem. 2009;284:16482–16491. doi: 10.1074/jbc.M808255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Steffen KK, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kujoth GC, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 76.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 77.Dillon LM, et al. Increased mitochondrial biogenesis in muscle improves aging phenotypes in the mtDNA mutator mouse. Hum Mol Genet. 2012;21:2288–2297. doi: 10.1093/hmg/dds049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dillin A, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 79.Butler JA, et al. A metabolic signature for long life in the Caenorhabditis elegans Mit mutants. Aging cell. 2013;12:130–138. doi: 10.1111/acel.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cannon JR, et al. A highly reproducible rotenone model of Parkinson’s disease. Neurobiology of disease. 2009;34:279–290. doi: 10.1016/j.nbd.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Valente EM, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 82.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nature reviews. Molecular cell biology. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith GA, et al. The search for genetic mouse models of prodromal Parkinson’s disease. Experimental neurology. 2012;237:267–273. doi: 10.1016/j.expneurol.2012.06.035. [DOI] [PubMed] [Google Scholar]
- 84.Tain LS, et al. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nature neuroscience. 2009;12:1129–1135. doi: 10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]