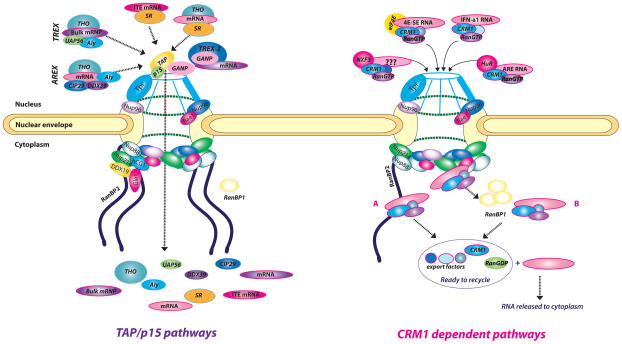
Figure 1. TAP1 and CRM-1 dependent mRNA export pathways are heterogenous demonstrating substantial plasticity.
As described in the text, factors associating with specific RNAs, often through USER codes, underlie formation of specific export mRNPs. The NPC dependent RNA export pathways are divided into two major subtypes, TAP and CRM1 dependent. Further subdivisions are indicated by the different composition of the nuclear mRNPs. For TAP and its cofactor NXT1/p15 dependent export, complexes that depend on Aly/REF (TREX and AREX), additionally on GANP (as a part of TREX-2 complex), and distinct complexes involving the SR proteins are shown (ITE mRNA represents intronless mRNAs). For the TAP pathways, release of cargoes is depicted using the DDX19/Gle1 model, which is hypothesized primarily from yeast data to be present in humans [9, 25]. Four subdivisions of the CRM1 pathway are similarly depicted by different nuclear mRNP complexes for HuR, eIF4E, IFN-α1 and NXF3. At the nuclear basket and cytoplasmic face, nucleoporins and co-factors described in the text are shown. The cytoplasmic side depicts only 4 fibrils, which is a simplification for presentation purposes (as there are known to be eight fibrils per NPC [8, 10]). For CRM1 dependent pathways, the RanGTP cycle is shown for cargo release, which can occur using RanBP2 (A) or RanBP1 (B). For the case of the eIF4E dependent mRNA export pathway, RanBP2 is depleted, RanBP1 is elevated and thus the RanBP1 release pathway is thought to predominate. Once cargoes are released, factors are recycled. Note the color difference of RanGTP (purple) and RanGDP (green). Many of these factors are dysregulated in cancer (see text).