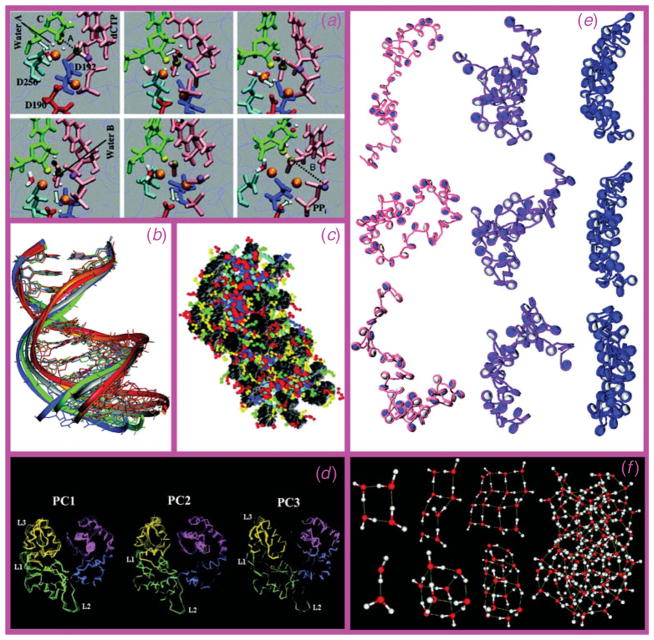
Fig. 1.
Examples of various modeling methods and applications. (a) QM/MM pathway (from top left to bottom right) of DNA repair enzyme polymerase β nucleotide incorporation (Radhakrishnan & Schlick, 2006); (b) superimposed MD configurations from a solvated dodecamer simulation ; (c) mesoscale oligonucleosome model with nucleosome cores in grey and tails and linker histones in color (see Arya & Schlick, 2009; Schlick, 2009a for details) ; (d) top three principal component motions in the thumb and finger subdomains of DNA polymerase β (Arora & Schlick, 2004); (e) representative Monte Carlo snapshots in the sampling of 48-unit oligonucleosomes at three different salt environments using the model shown in (c) ; and (f) minimized water clusters.