Abstract
Dielectrophoresis (DEP) can be used to noninvasively measure the dielectric state of the cell, and this data can be used to monitor cell health or apoptosis. In this study, we followed events associated with cytosine arabinoside (Ara-C)-induced apoptosis in NB4 cells using DEP analysis. Our data showed that the membrane capacitance of NB4 cells decreases from 9.42 to 7.63 mF/m2 in the first 2 hours following treatment with Ara-C, and that this decreased capacitance persists for >12 hours. Additionally, cytoplasmic conductivity decreases from 0.217 to 0.190 S/m within 2 hours of Ara-C treatment; this level is maintained for a short period of time before decreasing. We also investigated these events molecularly at the level of gene expression using microarray analysis and showed that the expression of genes related to membrane capacitance and cytoplasmic conductivity change dramatically as early as 2 hours post-Ara-C treatment, and further demonstrated a temporal relationship between the dielectric properties and key events in apoptosis. This study, integrating physical electrical properties of the cell membrane and cytoplasm with those of conductivity-related gene networks, provides new insights into the molecular mechanisms underlying the initiation of apoptosis, establishing a systematic foundation for DEP application in follow-up drug screening and development of medicines for treating leukemia.
Keywords: apoptosis, dielectrophoresis, gene expression profiling, leukemia
Introduction
Approximately 1200 new cases of acute promyelocytic leukemia (APL) are diagnosed in the United States each year (http://ghr.nlm.nih.gov/). The most widely-used anti-metabolite used to induce remission in children and adults with acute leukemia1 and for APL consolidation therapy to reduce the relapse rate2 is cytosine arabinoside (Ara-C). Ara-C is a potent killer of dividing cells by inducing cellular apoptosis mechanisms.
Apoptosis is an essential cell process for the ordered destruction of unwanted or damaged cells and can be defined as a gene-directed cellular self-destruction mechanism. Dysfunction of this mechanism is linked to the pathogenesis of cancer and other diseases3 whereby reduced apoptosis prolongs the persistence of transformed cancer cells. Thus, in vitro technologies for sensitive and rapid assessment of apoptotic events may be useful for understanding cellular events involving the apoptosis mechanism and for aiding disease treatment and anti-cancer drug development.
Many features can be used to distinguish normal cells from cells undergoing apoptosis, such as decreased mitochondrial transmembrane potential,4 externalization of phosphatidylserine (PS),5 changes in morphological features6,7 and changes in intracellular ion concentrations.8 Hence, various methods have been developed to detect apoptosis. The most commonly used method is in vitro measurement of fluorescence probes designed for specific labeling of relevant molecules in apoptotic cells, such as Annexin V and JC-1 assays.9–11 1 However, these approaches are typically invasive, resulting in cell death.
It has been long recognized that dielectrophoresis (DEP) can be used to determine the dielectric properties of a cell. DEP is noninvasive, does not require markers or labels, and has been developed for a broad range of applications. For instance, DEP can be used for cell characterization,12 cell isolation,13 cell separation,14 drug resistance monitoring,15 drug assessment,16 in vitro fertilization,17 and stem cell research,18,19 as well as for analyzing cells in different physiological states, particularly apoptosis.20–24 However, the exact molecular mechanisms that occur during apoptosis remain unclear, as are the events associated with Ara-C-induced apoptosis.
In this study, Ara-C-induced NB4 cell apoptosis was monitored using DEP analysis and validated using standard Annexin V and JC-1 assays. A 22 K human genome oligo array was also used to examine expression levels of genes associated with membrane capacitance and cytoplasmic conductivity in parallel with DEP measurements to comprehensively investigate molecular genetic events that occur during apoptosis.
Material and methods
Cell culture and induction of apoptosis
The NB4 cell line was obtained from Peking Union Medical College. The cells were maintained in RPMI 1640 media (Gibco, Grand Island NY, USA) supplemented with 10% (v/v) fetal bovine serum (FBS; PAA, Pasching, Austria), and 1% (v/v) penicillin/streptomycin (Invitrogen, Carlsbad CA, USA). The cells were grown at 37°C in a 5% CO2 atmosphere. Ara-C was purchased from Sigma-Aldrich (St Louis, MO, USA). To induce apoptosis, NB4 cells (at a density of 3 × 105 cells/mL) were treated with Ara-C at a final concentration of 1 × 10−5 M. Concentration was determined using CCK-8 assays (see Supplementary material). The incubation period included four time points: 2, 4, 6, and 12 hours.
Detection of apoptosis using Annexin V and JC-1 assays
Apoptosis was analyzed using flow cytometric analysis (FCA). Cells were cultured in 6-well plates, treated with 1 × 10−5 M Ara-C, collected at different time points, and centrifuged at 190 × g for 5 minutes. A rapid Annexin V-FITC apoptosis detection kit (KeyGEN Biotech, Nanjing, China) was used according to the manufacturer’s instructions. Analysis was performed on a BD FACSAria™ II system (BD Biosciences, San Jose, CA, USA). Cells incubated without Ara-C were analyzed in parallel as controls.
JC-1 was used to dye NB4 cells to visually detect early apoptosis. Similarly to the procedure for flow cytometric analysis, cells were collected and incubated in the dark in 0.5 mL incubation buffer (KeyGEN Biotech) with 1 μL JC-1 (2.5 mg/mL) at 37°C for 20 minutes. After incubation, the cells were washed twice with 1 × PBS and analyzed using fluorescence microscopy (DM-IRB; Leica, Wetzlar, Germany). To reduce the presence of dye particulates prior to incubation, the JC-1 solution was sonicated for 5 minutes, followed by centrifugation (1 minute at 9300 × g). The supernatant was used for dye loading.
Dielectrophoretic system setup and crossover frequency determination
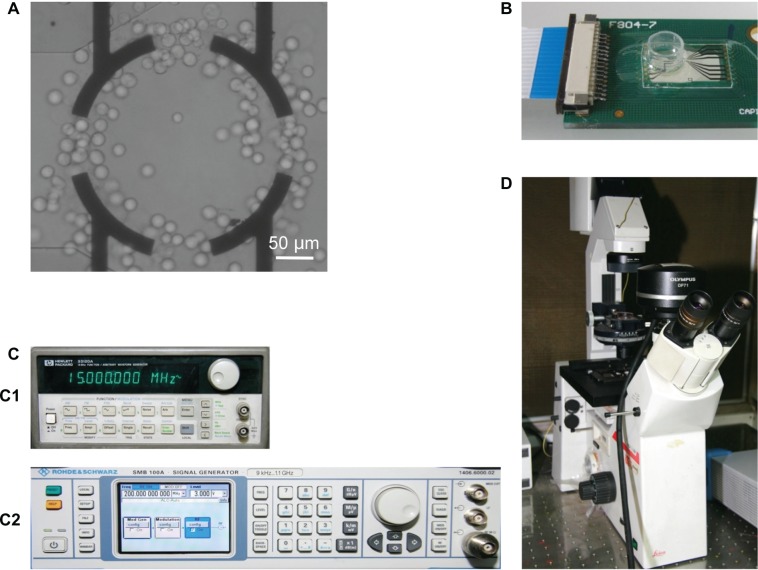
The principal components of the DEP system are described in Figure 1. The DEP chip was fabricated by patterning nonclosed ring gold electrodes onto the surface of a clean glass wafer using standard photolithography. As shown in Figure 1A, the width of the electrodes on the glass chip was 20 μm and the inner diameter of the ring was 200 μm. A 200-μL Eppendorf tube was cut and assembled on the chip to construct the DEP measurement chamber (Figure 1B).
Figure 1.
Principal components of the DEP assay system. (A) Nonclosed ring gold electrodes, with width of 20 μm and inner diameter of 200 μm. (B) The assembled chip with a reservoir modified from a 200-μL Eppendorf tube. (C) Signal generator. C1: HP33120A, C2: SMB100A. (D) CCD camera coupled to a fluorescence microscope.
Abbreviation: DEP, dielectrophoresis.
The sine signal applied to the chip was generated by a signal generator (HP33120A; Hewlett-Packard Santa Clara, CA, USA) in the range of 10–15 MHz or a signal generator (SMB100A; Rohde and Schwarz, Munich, Germany) in the range of 50–500 MHz at an applied voltage of 3 V (peak-to-peak). Measurements were observed through a CCD camera (DP-71; Olympus, Tokyo, Japan) coupled to a fluorescence microscope (DM-IRB; Leica).
Prior to DEP measurements, the cells were centrifuged at 190 × g for 5 min. The pellets were washed and resuspended in a prepared isotonic medium containing 8.5% (w/v) sucrose and 0.3% (w/v) dextrose buffer for which conductivity had been adjusted to 32.8 mS/m with RPMI 1640 media and a conductivity meter (DDSJ-308A; SPSIC Ltd Shanghai, China). Subsequently, the cells were added to the chip chamber using a micropipette, and the motion of the cells toward or away from the electrode edges due to the applied frequency was observed.
DEP measurements are made by determining crossover frequencies, at which most of the cells in the chamber experience a zero DEP-induced force and exhibit no movement (ie, corresponding to the transition between negative and positive DEP). The crossover frequency for ≥20 cells was measured over a time period of ≤20 minutes for each experiment. The frequency obtained was further used to determine the capacitance of the plasma membrane23 or cytoplasmic conductivity.25
Gene expression profiling and real-time quantitative polymerase chain reaction (PCR)
The 22 K human genomic oligo array (CapitalBio Corp, Beijing, China) contains 21,329 5′-amino-modified 70-mer probes of the Human Genome OligoSet (Version 2.1; Operon, Huntsville, AL, USA). Total RNA was isolated using the Trizol method (Invitrogen). Reverse transcription was performed using M-MLV (Takara Chemicals, Shiga, Japan). Microarray experiments were performed as described previously.26 After hybridization, microarrays were scanned using a LuxScan™ 10 K/A confocal scanner (CapitalBio), and data from the obtained images were extracted using LuxScan 3.0 software (CapitalBio). Raw data were normalized using the space- and intensity-dependent LOWESS program.27 For each test and control sample at the 2-, 4-, 6-, and 12-hour time points, hybridization experiments were performed using the dye-swap strategy. Only genes with consistent differential expression (both above a twofold change) in both dye-swap microarrays were selected as differentially expressed genes. The description of this microarray study follows the Minimum Information About a Microarray Experiment (MIAME) guidelines.28
A selected subset of differentially expressed genes was validated using real-time quantitative PCR employing an EvaGreen Real-time qPCR Core Reagent Kit (CapitalBio) and a Bio-Rad IQ™ 5 (Bio-Rad, Hercules, CA, USA). Target genes and a reference gene (glyceraldehyde 3-phosphate dehydrogenase [GAPDH]) were amplified in parallel. Primer sequences are shown in Supplementary Table S1. The results were analyzed using Bio-Rad IQ™ 5 (software version 2.1; Bio-Rad). PCR amplification products were analyzed using melting curve analysis and 1.5% agarose gel electrophoresis. The formula was employed to calculate the relative gene expression ratio of a target gene, where E is the real-time PCR efficiency and CP is the real-time PCR crossing point.29 All reactions were performed in triplicate.
Detection of morphology changes using scanning electron microscopy (SEM) and indirect immunofluorescence (IF) microscopy
After centrifugation at 190 × g for 5 minutes followed by resuspension, the cells were fixed with 2.5% glutaraldehyde at room temperature for 1 hour, washed in 1 × PBS, and exposed to 1% osmium tetroxide for another hour. The cells were dehydrated by sequential immersion in 30, 50, 70, 80, 90, and 2 × 100% ethanol, followed by a 50/50 ethanol/tert-Butanol solution, and then 100% tert-Butanol. The cells were freeze-dried (ES-2030; Hitachi, Tokyo, Japan) for 4 h, coated with a thin gold layer using an ion sputter (E-1010; Hitachi), and finally attached to the microscope supports using silver glue. The cells were imaged at 15 kV by SEM (Quanta 200; FEI, OR, USA).
For IF analysis, cells were collected washed and plated on glass coverslips. Next, the cells were fixed in 4% paraformaldehyde for 30 min and permeabilized using 0.2% Triton X-100 in 1 × PBS. After blocking with 1% (w/v) bovine serum albumin (BSA) in 1 × PBS, the cells were incubated with rabbit KIF20A polyclonal antibody (Abnova, Taipei, Taiwan) or rabbit y-tubulin polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4°C overnight. Subsequently, the cells were washed three times with 1 × PBS and stained with DyLight-labeled goat anti-rabbit IgG (Jackson, West Grove, PA, USA) at room temperature for 4 hours. Nuclei were stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI); Vector Labs, Burlingame, CA, USA). The cells were then examined and photographed using a laser scanning confocal microscope (LSM710; Carl Zeiss, Oberkochen, Germany). The excitation wavelength was 488 nm for DyLight and 405 nm for DAPI. Images were merged using ZEN software (Carl Zeiss).
Measurements of intracellular calcium, sodium, and potassium using flow cytometry
Intracellular calcium ion concentration was measured using Fluo4-AM (Dojindo, Kumamoto, Japan) and 20% pluronic F-127 dissolved in DMSO to make a 1 mM stock solution. Immediately before Fluo 4-AM loading, the cells were gently washed with HEPES-buffered saline (10 mM HEPES, 137 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 10 mM D-glucose, pH 7.35) and then incubated in Fluo 4-AM working solution (5 μM Fluo 4-AM and 0.05% pluronic F-127 dissolved in HEPES buffer) for 30 minutes at 37°C in a cell incubator. Next, the cells were washed with HEPES-buffered saline to remove extracellular Fluo 4-AM and incubated for an additional 20 minutes to allow deesterification of Fluo 4-AM into its active-dye form Fluo 4. Analysis was performed on a BD FACSCalibur™ system (BD Biosciences) at an excitation of 488 nm.
For intracellular sodium and potassium ion measurements, 2 μL of 2.5 mM SBFI-AM (Sigma-Aldrich) and PBFI-AM (Santa Cruz Biotechnology) stock solutions were individually added to 1 mL of cells at a final concentration of 5 μM 1 hour prior to conducting the experiment. Incubation was continued at 37°C in a 5% CO2 atmosphere. Immediately before FCA, propidium iodide (PI; Sigma-Aldrich) was added at a final concentration of 10 μg/mL. Approximately 1 × 104 cells were analyzed by sequential excitation of the cells at 355 nm for SBFI-AM or PBFI-AM and 488 nm for PI, using a BD™ LSR II Flow Cytometer System (BD Biosciences).
Results and discussion
Detecting apoptosis with Annexin V and JC-1 assays
Apoptotic cells display significant externalization of PS, which can be validated using the Annexin V assay. As shown in Supplementary Figure S2, a series of the Annexin V assays based on flow cytometry was performed at different time points. Our results demonstrate that the proportion of Annexin V-positive cells at 2 hours posttreatment was not significantly different compared to control cells, and up to 4 hours posttreatment, the proportion of the positive cells displayed a clear difference (Figure 2A). Among the entire cell population, this proportion increased from 4.51% ± 0.44% at 2 hours to 7.19% ± 0.26% at 4 hours, 12.42% ± 0.68% at 6 hours, and 21.68% + 0.45% at 12 hours.
Figure 2.
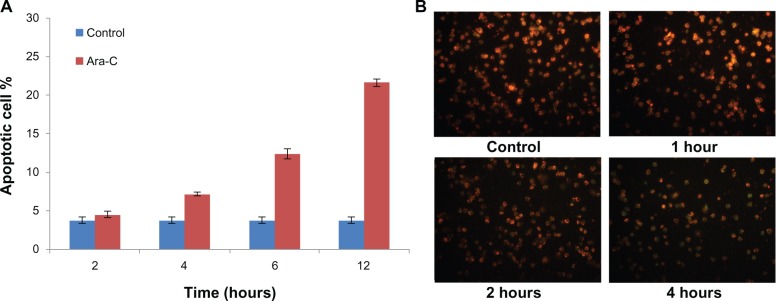
Detection of apoptosis by Annexin V and JC-1 assays. (A) Percentage of apoptotic cells after different incubation periods with Ara-C as measured using FCA and Annexin V/PI assays. (B) Viable cells display greenish orange fluorescence and apoptotic cells display green fluorescence.
Note: Our results suggest that NB4 cell apoptosis initiated by Ara-C can be detected at 2 h post-treatment using the JC-1 assay.
Abbreviations: Ara-C, cytosine arabinoside; FCA, flow cytometric analysis.
To ensure that apoptosis initiated by Ara-C could be detected as early as possible (ie, at 2 hours posttreatment), assays based on the mitochondria membrane potential (ΔΨm) were performed. A typical early characteristic of apoptosis is permeabilization of the mitochondrial membrane followed by dissipation of ΔΨm, which is measured using the JC-1 assay10 As shown in Figure 2B, cells displaying greenish orange fluorescence were viable, while cells displaying green fluorescence were apoptotic. After 2 hours of Ara-C treatment, the proportion of apoptotic cells increased. This indicates that NB4 cell apoptosis initiated by Ara-C can be detected as early as 2 hours posttreatment using the JC-1 assay, during which time the mitochondrial permeability transition pore opens and the corresponding ΔΨm is reduced.
DEP analysis during Ara-C-induced apoptosis
DEP analysis is an assay based on measuring changes in dielectric properties (ie, membrane capacitance and cytoplasmic conductivity) that has been applied for monitoring cell apoptosis.20–24 Currently, there are several theoretical models used to extract cell dielectric properties.30,31 In this study, we examined the variation in cell dielectric property trends during cell apoptosis. We applied a simple model for measuring crossover frequencies in an isotonic medium with a specific conductivity (32.8 mS/m).23,25 DEP analysis reveals two types of crossover frequencies, the lower and upper crossover frequencies, which are indicated by ƒx1 and ƒx2, respectively. ƒx1 provides a direct measurement of membrane capacitance, while fx2 provides a direct measurement of cytoplasmic conductivity.25 As shown in Figure 3A and B, crossover frequency of the cells at 2 h post-exposure to Ara-C displayed distinct differences with that obtained from control cells; moreover, ƒx1 continually increased from 96 ± 4.73 to 354 ± 6.11 kHz, while fx2 decreased from 301 ± 7.09 to 165 ± 7.78 MHz over the 12-hour time course. Additionally, cell diameter decreased from 16.30 ± 0.35 to 13.81± 0.62 μm (Figure 3C). As shown in Table 1, membrane capacitance (Cm) decreased from 9.42 ± 0.47 to 3.03 ± 0.05 mF/m2 over the 12-hour time course based on the formula of Pethig and Talary ,23 where r and σs represent the cell radius and conductivity of the surrounding solution (32.8 mS/m), respectively. The decreasing trend of membrane capacitance observed in our experiments is consistent with the observations of previous studies.23,24 Similarly, we calculated that cytoplasmic conductivity (σ) decreased from 0.217 ± 0.005 to 0.120 ± 0.006 S/m over the 12-hour time course based on the equation ,25 where ɛs indicates the relative permittivity of the surrounding solution is a constant, about 81. This generally agrees with previous studies examining Jurkat T-cells undergoing etoposide-induced apoptosis.23 Compared to the Annexin V and JC-1 assays, the DEP assay can be used to monitor apoptosis noninvasively without the use of added markers or labels.
Figure 3.
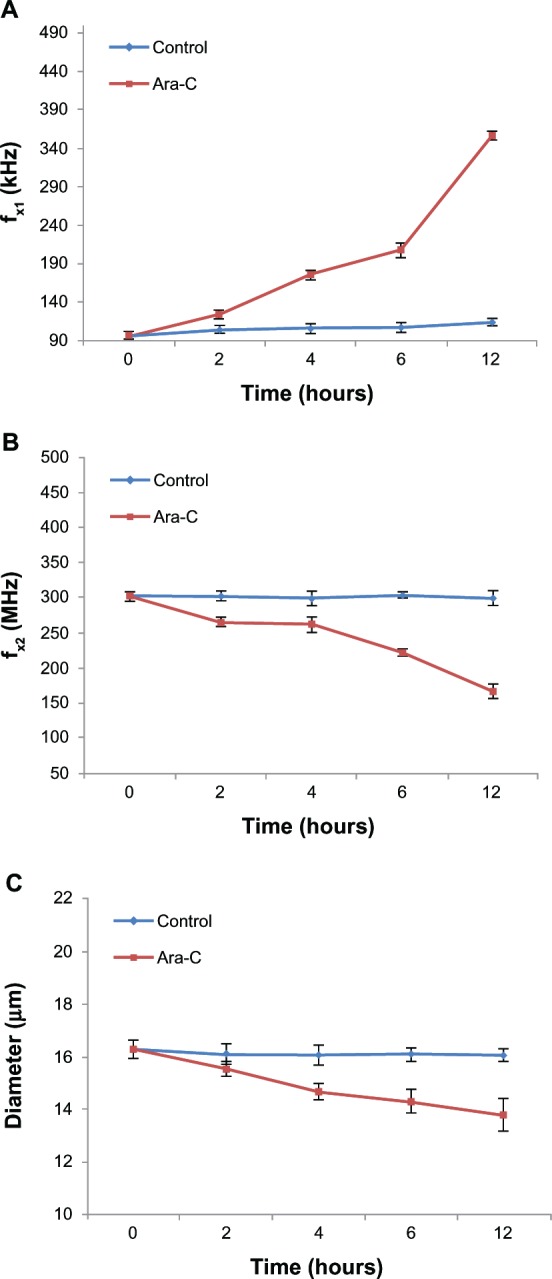
Monitoring of Ara-C induced apoptosis by DEP analysis. (A and B) Distinct differences in crossover frequency occur between control and the apoptotic NB4 cells as early as 2 h post-exposure to Ara-C. ƒx1 increased from 96 ± 4.73 to 354 ± 6.11 KHz, while ƒx2 decreased from 301 ± 7.09 to 165 ± 7.78 MHz over the 12-h time course. (C) Cell diameter decreased from 16.30 ± 0.35 to 13.81 ± 0.62 μm in apoptotic cells.
Note: No significant change was observed for control cells.
Abbreviations: Ara-C, cytosine arabinoside; DEP, dielectrophoresis.
Table 1.
Changes in membrane capacitance (Cm) or cytoplasmic conductivity (σcyto) during Ara-C-induced apoptosis
| Time (hours) | Cm(mF/m2) | σcyto (S/m) |
|---|---|---|
| 0 (control) | 9.42 ± 0.47 | 0.217 ± 0.005 |
| 2 | 7.63 ± 0.25 | 0.190 ± 0.005 |
| 4 | 5.75 ± 0.21 | 0.188 ± 0.004 |
| 6 | 4.93 ± 0.20 | 0.160 ± 0.004 |
| 12 | 3.03 ± 0.05 | 0.120 ± 0.006 |
Gene expression profiling during Ara-C-induced apoptosis
To better understand the molecular mechanism underlying apoptosis using DEP monitoring, we examined mRNA changes in NB4 cells after Ara-C treatment over a 12-hour time course using a human whole genome oligo array. A total of 9, 37, 42, and 117 genes displayed altered expression levels at 2, 4, 6, and 12 hours, respectively, as shown in Supplementary Table S2. A total of 152 genes showing ≥2.0-fold change at least once for all four time points (Supplementary Table S3) were selected for gene ontology (GO) analysis and cluster analysis (Supplementary Figure S3).
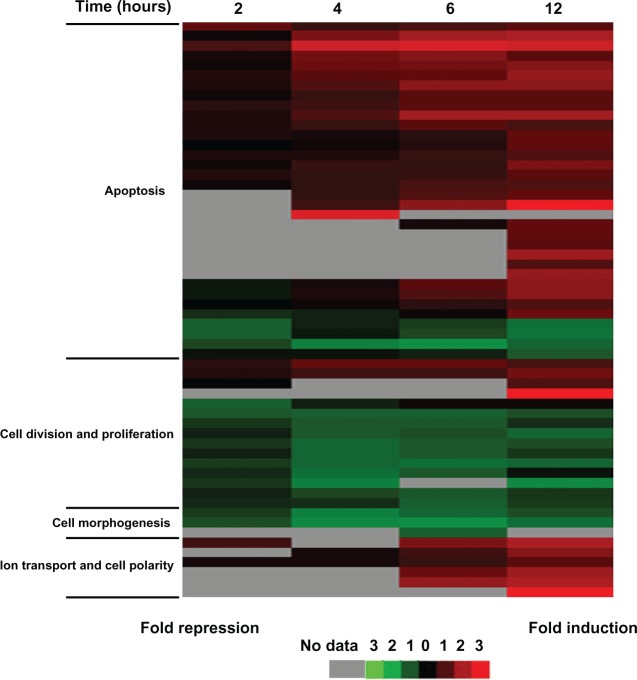
GO analysis revealed that differentially expressed genes can be grouped into five main categories: apoptosis, cell division and proliferation, cell morphogenesis, ion transport and cell polarity, and others. Additionally, cluster analysis confirmed the expression level of some known apoptosis-related genes changed after Ara-C treatment, including the up-regulated pro-apoptosis genes SESN2, E2F2, DDIT4, TNF, GADD45A, and PMAIPI and the downregulated antiapoptosis genes HSPA1A, HSPA1B, and ARHGDIA (Figure 4, Supplementary Figure S4(A)). In contrast, the expression level of nearly all genes involved in cell division and proliferation was downregulated (eg, C20orf129 and IRS2) (Figure 4, Supplementary Figure S4(B)). Notably, the expression level of some genes involved in cell morphogenesis or ion transport changed as early as 2 hours post-Ara-C treatment, which may correlate with the measured alteration in membrane capacitance or alteration in cytoplasmic conductivity (Figure 4, Supplementary Figure S4(C and D)). Furthermore, we analyzed expression changes in terms of transcriptional regulation of defined gene networks and pathways and present a model of the dynamic and temporal network of pathway activities related to Ara-C-induced apoptosis (Supplementary Figure S5), which may have implications for identifying potential new effectors useful for treating APL.
Figure 4.
Relationship of molecular events to the time course of Ara-C-induced apoptosis.
Note: Cluster image of gene expression data of the four main categories: apoptosis, cell division and proliferation, cell morphogenesis, and ion transport and cell polarity.
Abbreviation: Ara-C, cytosine arabinoside.
The original data from this study are available at the Gene Expression Omnibus (see http://www.ncbi.nlm.nih.gov/geo); the accession number is GSE32992. Results of real-time quantitative PCR analysis for all selected genes were in agreement with microarray data (Supplementary Figure S4).
Systematic analysis of changes in dielectric properties
Because DEP analysis is noninvasive, nondestructive, and rapid it has been widely employed for cell apoptosis studies.20–24 However, little is known regarding the temporal relationship between dielectric properties measured using DEP analysis and key events in apoptosis. Here, we used DEP analysis to systematically investigate potential mechanisms responsible at the gene level and at the cellular level for changes in membrane capacitance and cytoplasmic conductivity occurring during apoptosis.
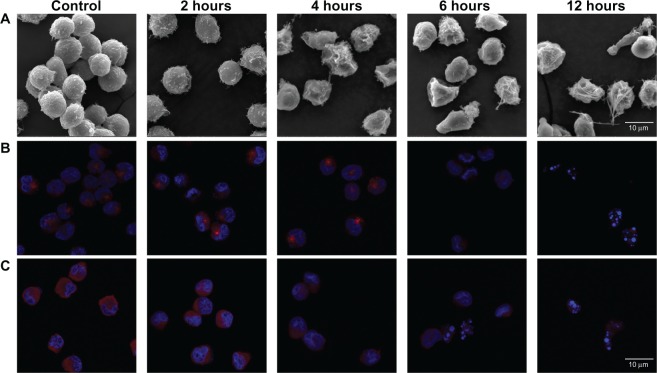
As described above, membrane capacitance decreased from 9.42 ± 0.47 to 3.03 ± 0.05 mF/m2 over 12 hours. Cell membrane capacitance largely depends on plasma membrane surface morphology, such as microvilli, ruffles, folds, and blebs;32–35 cells treated with Ara-C, as well as control cells, were imaged using SEM (Figure 5A). Control cells were covered with numerous microvilli, which appeared to be homogeneous both in size (~16 μm diameter) and surface morphology. However, treated cells exhibited alterations in their plasma membrane morphology. After 2 hours of treatment with Ara-C, the number of microvilli slightly decreased, and the membrane surface became smoother. After a treatment time of ≥4 hours, cell shrinkage and bleb formation were observed. Membrane surface morphology is closely associated with the cytoskeleton36 and thus can be monitored by observing changes in microtubule-related proteins during apoptosis.37,38 As shown in Figure 5B, the average cell fluorescence intensity (FI) related to γ-tubulin from the IF assay gradually decreased over the 12-hour time course, indicating that in cells treated with Ara-C, micro-tubules were disrupted and collapsed, resulting in obvious changes to the cytoskeleton as observed by others.37 Changes in surface morphology involving the cytoskeleton effectively decreased the surface area of the cell membrane and, hence, decreased cell membrane capacitance.
Figure 5.
Changes in morphological features of cells over the 12-h time course of Ara-C induced apoptosis. (A) SEM images of NB4 cells after treatment for 0 (control), 2, 4, 6, and 12 h with Ara-C. The cells displayed progressive changes in membrane morphology as apoptosis developed. (B and C) Merged confocal scanning laser microscopy images represent the changes in microtubule-related proteins over the 12-h time course: (B) γ-tubulin and (C) KIF20A.
Note: Cell nuclei are stained with DAPI (blue).
Abbreviations: Ara-C, cytosine arabinoside, SEM, scanning electron microscopy; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride.
We examined the expression level of microtubule-related genes, such as kinesin family member 20A (KIF20A)39 and centromere-associated protein E (CENPE),40,41 and found that these genes were downregulated over the 12-hour time course. More precisely, expression of KIF20A and CENPE decreased to a minimum at 4 hours posttreatment and then remained low for the remainder of the time course (Supplementary Figure S4(C)). Accordingly, the decline in average cell fluorescence intensity related to KIF20A in the IF assay reflects the decreasing KIF20A protein quantity and activity (Figure 5C), which was similar to γ-tubulin. Because of the close association between membrane surface morphology and KIF20A, a cytoskeletal component,36 the decline in KIF20A expression may induce structural changes to the cell membrane (Figure 5A), further decreasing cell membrane capacitance.33,34 Subsequently, an increase in ƒx1 was observed as a consequence of these changes.
Additionally, cytoplasmic conductivity decreased from 0.217 ± 0.005 to 0.120 ± 0.006 S/m. σcyto is another important cell dielectric property which is, to a large extent, determined by ion mobility and concentration in the cytoplasm.42 Our gene expression profiling results demonstrated that expression of ion transport-related genes, such as P2X purinoceptor 4 (P2RX4; associated with calcium and sodium transport)43,44 and potassium channel tetramerisation domain containing 9 (KCTD9; associated with potassium transport),45,46 was up-regulated over the 12-hour time course (Supplementary Figure S4(D)). Moreover, changes in intracellular calcium, sodium, and potassium concentration were measured using fluorescence probe-based flow cytometry, and all changes in cytoplasmic conductivity over the 12-hour time course were estimated using the equation of Bard and Faulkner.47 The experimental data revealed an increase in intracellular calcium and sodium, as well as a loss of intracellular potassium, during apoptosis (Supplementary Figure S6(A–C)). Figure 6 shows the relative dynamic and temporal changes in intracellular ions and cytoplasmic conductivity over the 12-hour time course. Intracellular calcium and sodium increased to >1.5 and 1.2 times their normal levels, respectively, while intracellular potassium level was reduced to <0.80 times its normal level over the 12-hour time course (Figure 6A). A decrease in intracellular potassium ultimately induces a loss of cytoplasmic conductivity because it is a highly abundant intracellular ion,48 even when intracellular calcium and sodium levels increase. Estimated relative changes of average cytoplasmic conductivity based on ion levels decreased significantly at 2 hours post-Ara-C treatment and changed minimally thereafter. T DEP analysis showed nearly the same results over the 6-hour time course. However, the difference became significant at the 12-hour time point (Figure 6B). One possible reason for this difference may be that the ions contributing to cytoplasmic conductivity were limited. Based on gene expression profiling, we found that the expression level of chloride intracellular channel 4 (CLIC4; associated with chloride transport)49,50 was significantly up-regulated at the 12-hour time point (Supplementary Table S3), indicating that an apparent change of intracellular chloride ions may have occurred, further affecting overall cytoplasmic conductivity. As a label-free method for measuring overall cytoplasmic conductivity, DEP analysis may be a better choice as a simple and quick monitoring method.
Figure 6.
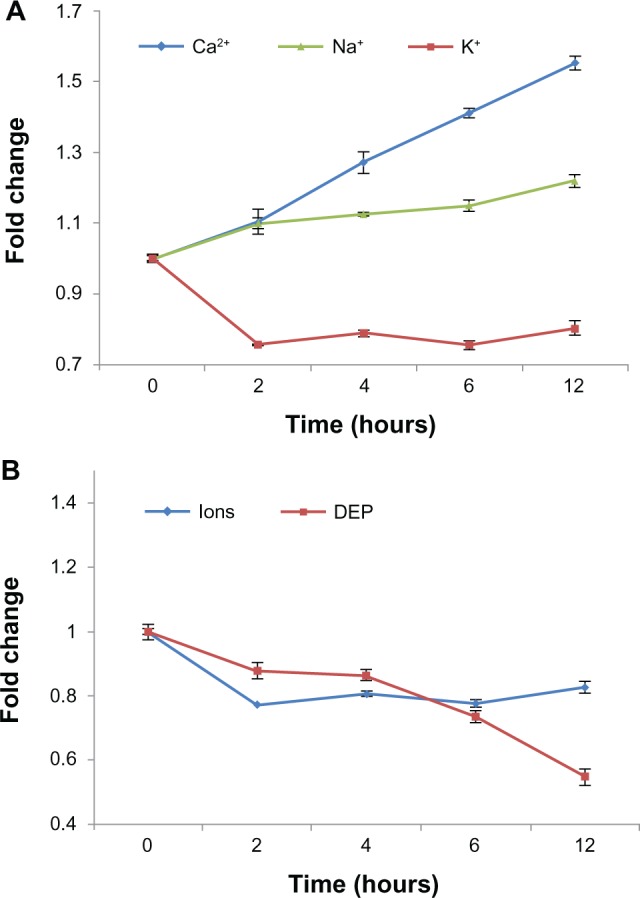
Relative dynamic and temporal changes in intracellular ion levels and cytoplasmic conductivity over the 12-h time course. (A) Fold changes in intracellular calcium, sodium, and potassium from the 0- to 12-hour time points during Ara-C-induced apoptosis. The data represent averages of three independent experiments. (B) Estimated changes in average cytoplasmic conductivity based on changes in intracellular ions levels (blue) or DEP analysis (red) over the 12-h time course.
Abbreviations: Ara-C, cytosine arabinoside; DEP, dielectrophoresis.
Conclusion
In this study, we used DEP analysis to investigate the mechanisms responsible for apoptosis from the gene and cell levels by performing a series of biological assays (gene expression profiling, SEM, IF, and FCA). At the gene level, our studies indicate that expression levels of KIF20A and CENPE, which are related to membrane capacitance, are down-regulated, while expression levels of P2RX4 and KCTD9, which are related to cytoplasmic conductivity, are up-regulated as early as 2 hours post-Ara-C treatment. Accordingly, at the cellular level, the FI related to γ-tubulin and KIF20A gradually decreased, and obvious structural changes to the cell membrane were observed, leading to the loss of membrane capacitance by decreasing the effective surface area of the cell membrane. Additionally, a decrease in intracellular potassium concentration induces a loss of cytoplasmic conductivity, even though the intracellular calcium and sodium levels increase. All of these changes in cell membrane capacitance and cytoplasmic conductivity make apoptotic cells amenable for characterization using DEP analysis, which may be used to help physicians detect apoptosis earlier. As this technology develops, individually tailored and more personalized patient treatment will be possible.
Supplementary material
Methods
Determining Ara-C concentration The appropriate working concentration of Ara-C was evaluated using Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) assays. Cells were plated at a density of 1 × 105 cells/mL in a 96-well plate. Incubation times and concentrations of Ara-C are shown with the results. After incubation, cell viability was determined using CCK-8 according to the manufacturer’s protocol. Absorbance at 450 and 630 nm was read using a Microplate Reader (Bio-Rad). The experiment was repeated in triplicate.
Results and discussion
As shown in Supplementary Figure S1, several different concentrations were used to test the effect of Ara-C on cell viability at various time points. IC50 (defined as the concentration of drug that reduces cell viability by 50% relative to untreated controls) values were calculated to be approximately 10−4 or 10−5 M for 12 h. Combined with the results of a previous report,51 a concentration of 10−5 M Ara-C was selected to induce cell apoptosis.
Inhibition of growth of NB4 cells by Ara-C.
Note: Relative viabilities were determined using CCK-8 assays as described in the experimental section and the results were compared with untreated (control) cells.
Abbreviations: Ara-C, cytosine arabinoside; CCK-8, Cell Counting Kit-8.
Example of FCA of a single cell apoptosis experiment.
Note: Five time points are shown for FCA, 0, 2, 4, 6, and 12 hours after addition of 10−5 M Ara-C; PI+/FITC+ indicates deeply apoptotic or dead cells, while PI−/FITC+ indicates apoptotic cells.
Abbreviations: Ara-C, cytosine arabinoside; FCA, flow cytometric analysis; h, hours.
Gene expression profiling using microarray. (A) Gene ontology image of differentially expressed genes. (B) Cluster image of gene expression profiles to Ara-C-induced apoptosis from 0- to 12-hour time points.
Abbreviations: Ara-C, cytosine arabinoside; h, hours.
Real-time quantitative PCR analysis of the expression of selected genes. The results of real-time quantitative PCR analysis were similar to those microarray gene expression data. (A) Apoptosis-related genes. (B) Cell division- and proliferation-related genes. (C) Microtubule-related genes. (D) Ion transport-related genes.
Abbreviations: PCR, polymerase chain reaction.
Systematic transcription regulation pathway map of the apoptosis time course.
Notes: Solid dark lines represent previously known pathways; dashed dark lines represent undefined pathways. Red rectangles represent induced genes with fold changes ≥ 1.5; green rectangles represent repressed genes with fold changes ≥ 0.67. Blue ovals represent hypothetical activated TFs. Further TF ELISA experiments are needed to determine whether p53 or NF-κB activation is involved in Ara-C-induced apoptosis of NB4 cells. All elements are approximately arranged relative to the time axis.
Abbreviations: Ara-C, cytosine arabinoside; NF-κB, nuclear factor κB; TF, transcription factor.
Changes in intracellular calcium, sodium, and potassium over the 12-h time course measured using FCA. (A) An increase in Fluo 4-AM (Ca2+) fluorescence from control cells indicates an increase in intracellular calcium. (B) An increase in SBFI-AM (Na+) fluorescence from control cells indicates an increase in intracellular sodium. (C) A decrease in PBFI-AM (K+) fluorescence from control cells indicates a loss of intracellular potassium.
Notes: The data are shown as histograms which are representative of a single experiment.
Abbreviations: FCA, flow cytometric analysis h, hours.
Sequences of primer pairs used for real-time quantitative PCR
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| SESN2 | AGATGGAGAGCCGCTTTGAGCT | CCGAGTGAAGTCCTCATATCCG |
| E2F2 | AGCTGGAACCGAGAGAACATG | ACACGACCAGGCGAAACC |
| TNF | GGCGTGGAGCTGAGAGATAAC | GGTGTGGGTGAGGAGCACAT |
| HSPA1A | ACCTTCGACGTGTCCATCCTGA | TCCTCCACGAAGTGGTTCACCA |
| HSPA1B | ACCTTCGACGTGTCCATCCTGA | TCCTCCACGAAGTGGTTCACCA |
| C20orf129 | GCGTGGCAACAGGCTCCTACA | GGCATACAGGATTCGGAACTCC |
| IRS2 | CCTGCCCCCTGCCAACACCT | TGTGACATCCTGGTGATAAAGCC |
| DDIT4 | AGGAAGCTCATTGAGTTGTG | GGTACATGCTACACACACAT |
| KIF20A | GTACCAACCAGGAAAATCAG | TGTCTGAGTATTGCATCCTG |
| CENPE | GGAGAAAGATGACCTACAGAGGC | AGTTCCTCTTCAGTTTCCAGGTG |
| P2RX4 | GTGGCGGATTATGTGATACCAGC | CACACAGTGGTCGCATCTGGAA |
| KCTD9 | GCCGCTGTAATCTTGCACATGC | CAGTTTCAGGGATGCTCCTTCTG |
| GAPDH | CGCTCTCTGCTCCTCCTGTT | CCATGGTGTCTGAGCGATGT |
Abbreviation: PCR, polymerase chain reaction.
Number of differentially expressed genes
| Time (hours) | Up | Down | Total |
|---|---|---|---|
| 2 | 5 | 4 | 9 |
| 4 | 15 | 22 | 37 |
| 6 | 24 | 18 | 42 |
| 12 | 95 | 22 | 117 |
Total of 152 genes with ≥2.0-fold change at least once for all four time points
| Oligo_id | Gene | Ratio
|
|||
|---|---|---|---|---|---|
| 2 hours | 4 hours | 6 hours | 12 hours | ||
| H200015775 | TNF | 2.2768 | 1.6131 | 1.8894 | 2.1108 |
| H200019847 | DDIT3 | 1.1387 | 2.8434 | 3.7666 | 4.1329 |
| H200011743 | DDIT4 | 1.865 | 5.208 | 5.8026 | 5.2344 |
| H200001568 | SESN2 | 1.1782 | 2.5341 | 2.9756 | 2.1333 |
| H200008354 | TXNIP | 1.1029 | 2.4659 | 2.6146 | 3.029 |
| H200015433 | E2F2 | 1.3478 | 2.0973 | 2.3398 | 3.3365 |
| H200000027 | PMAIP1 | 1.2504 | 1.9617 | 3.1681 | 3.0868 |
| H200006759 | GADD45A | 1.11 | 1.7016 | 2.1676 | 2.1329 |
| H200012407 | CASP8AP2 | 1.4683 | 1.7928 | 2.1279 | 2.1613 |
| H200003690 | TRIB3 | 1.2974 | 1.9831 | 3.8015 | 3.8056 |
| H200006040 | CHAF1B | 1.2458 | 1.6792 | 2.2018 | 1.8877 |
| H200005977 | PRNP | 1.2409 | 1.2069 | 1.4649 | 2.3403 |
| H200006833 | NFKBIA | 1.0624 | 1.1148 | 1.3696 | 2.2215 |
| H200006782 | IRF1 | 1.2729 | 1.2868 | 1.6145 | 2.0059 |
| H200000124 | ATF3 | 1.1052 | 1.6494 | 1.559 | 2.7846 |
| H200011520 | NAB1 | 1.3486 | 1.4977 | 1.6588 | 2.0771 |
| H200010637 | E2F8 | 1.1142 | 1.5773 | 1.8893 | 2.0674 |
| H200004695 | REL | 1.521 | 1.9931 | 2.2944 | |
| H200015131 | TNFAIP3 | 1.7875 | 3.1083 | 10.82 | |
| H200019156 | EGR1 | 5.9961 | |||
| H200003705 | RASSF1 | 1.2268 | 2.3082 | ||
| H200003433 | MOAP1 | 2.2819 | |||
| H200006172 | ASNS | 2.1505 | |||
| H200012696 | ZNF624 | 3.6537 | |||
| H200015127 | PRKCE | 2.055 | |||
| H200008824 | KLF6 | 3.2992 | |||
| H200006354 | BTG1 | 0.8407 | 1.1771 | 2.1741 | 3.1005 |
| H200006664 | NFIL3 | 0.8679 | 1.2632 | 1.9841 | 3.1347 |
| H200005485 | PAK1 | 0.9977 | 1.1359 | 1.4839 | 2.0311 |
| H200008426 | DUSP6 | 0.6807 | 0.7826 | 1.0939 | 2.3816 |
| H200001719 | HSPA1A | 0.4533 | 0.8086 | 0.6038 | 0.4123 |
| H200017080 | HSPA1B | 0.4746 | 0.8206 | 0.5659 | 0.3964 |
| H200005758 | EIF4EBP1 | 0.5687 | 0.3518 | 0.3128 | 0.4273 |
| H200007392 | ARHGDIA | 0.9244 | 0.8883 | 0.7877 | 0.4844 |
| H200001819 | CCNE1 | 1.4023 | 2.2679 | 2.2377 | 1.8367 |
| H200006902 | TIEG/KLF10 | 1.3368 | 1.8159 | 1.7091 | 2.5303 |
| H200000106 | ADM | 1.0576 | 2.0229 | ||
| H200000570 | CXCL10 | 10.3587 | |||
| H200013414 | IRS2 | 0.4683 | 0.7933 | 1.121 | 1.0842 |
| H200005718 | C20orf129/FAM83D | 0.491 | 0.4594 | 0.4546 | 0.5191 |
| H200001767 | TPX2 | 0.6455 | 0.4921 | 0.5083 | 0.6906 |
| H200006436 | DLG7/DLGAP5 | 0.765 | 0.4849 | 0.5174 | 0.445 |
| H200010905 | BUB1 | 0.6302 | 0.448 | 0.4873 | 0.5465 |
| H200012276 | ASPM | 0.7851 | 0.4425 | 0.5062 | 0.6396 |
| H200003347 | CCNB1 | 0.5973 | 0.425 | 0.4063 | 0.4447 |
| H200005304 | KNTC2/NDC80 | 0.716 | 0.398 | 0.514 | 0.8752 |
| H200006423 | PLK1 | 0.6491 | 0.35 | 0.334 | |
| H200004906 | CDCA8 | 0.7698 | 0.563 | 0.4997 | 0.6453 |
| H200005764 | ZNF339/OVOL2 | 0.7392 | 0.6969 | 0.4721 | 0.6271 |
| H200006136 | CENPE | 0.6114 | 0.3547 | 0.4507 | 0.5201 |
| H200005853 | KIF20A | 0.5375 | 0.3259 | 0.31 | 0.411 |
| H200012386 | ECT2 | 0.4648 | |||
| H200005775 | KCTD9 | 1.7487 | 2.7477 | 4.1513 | |
| H200018987 | P2RX4 | 1.1764 | 1.7371 | 2.8761 | |
| H200003474 | CLIC4 | 1.181 | 1.1757 | 1.6314 | 2.1938 |
| H200005870 | CCL3 | 2.417 | 3.5841 | ||
| H200006175 | CCL4 | 3.4727 | 3.9945 | ||
| H200003915 | TNFAIP6 | 11.9615 | |||
| H200020490 | 2.1693 | 2.0409 | 1.4841 | 1.6363 | |
| H200006467 | HMGCS1 | 2.1238 | 1.5023 | 1.1664 | 1.0935 |
| H200005214 | INSIG1 | 2.8237 | 1.373 | 1.1014 | 1.0765 |
| H200004003 | CCNE2 | 1.6662 | 2.6628 | 3.8329 | 3.4481 |
| H200002187 | TIPARP | 1.1651 | 2.0555 | 2.1295 | 2.3472 |
| H200017137 | CCL3L1 | 1.3932 | 1.2863 | 2.057 | 5.6597 |
| H200006526 | ATP2B1 | 1.141 | 1.0957 | 1.6069 | 2.9833 |
| H200002418 | FOXO3A | 1.1284 | 1.1058 | 1.5603 | 2.667 |
| H200001281 | HECA | 1.0933 | 1.1625 | 1.6718 | 2.8152 |
| H200005834 | 1.105 | 1.177 | 1.2049 | 2.2256 | |
| H200004109 | RSN | 1.0981 | 1.2093 | 1.6478 | 2.2426 |
| H200002137 | KIAA1147 | 1.1165 | 1.2148 | 1.4234 | 2.3482 |
| H200002754 | N4BP2 | 1.3414 | 1.2147 | 1.5133 | 2.1562 |
| H200014202 | RGS16 | 1.0903 | 1.2847 | 1.6735 | 3.4525 |
| H200011487 | CCNB1IP1 | 1.1918 | 1.2945 | 1.5251 | 2.2965 |
| H200012358 | MAP1LC3B | 1.0403 | 1.3294 | 1.9677 | 2.3889 |
| H200009957 | 1.1546 | 1.4168 | 1.7351 | 2.4842 | |
| H200011669 | C6orf48 | 1.1391 | 1.5785 | 1.4732 | 2.7148 |
| H200006727 | MMD2 | 1.764 | 1.6883 | 1.8576 | 2.657 |
| H200006646 | CD83 | 1.4052 | 1.7151 | 1.8771 | 2.8336 |
| H200002948 | ATP2B1 | 1.1653 | 0.9668 | 1.2938 | 2.2311 |
| H200007999 | GBP7 | 1.1948 | 0.9544 | 1.0879 | 2.2038 |
| H200007904 | PTPRC | 1.1887 | 0.8226 | 0.9554 | 2.1509 |
| H200006209 | C1orf56/MLLT11 | 1.2761 | 0.93 | 0.9883 | 2.2463 |
| H200017557 | OPN3 | 1.3918 | 1.1323 | 0.7939 | 0.4675 |
| H200009444 | 1.0565 | 1.0179 | 0.9952 | 0.4823 | |
| H200006408 | MAT2A | 1.1349 | 1.0642 | 0.6771 | 0.4931 |
| H200000511 | TBXAS1 | 1.0995 | 0.9085 | 0.7469 | 0.4983 |
| H200006231 | KIAA0182 | 1.1452 | 0.7109 | 0.6425 | 0.4688 |
| H200012473 | PCF11 | 1.1002 | 0.8868 | 0.6982 | 0.4149 |
| H200006260 | 1.1805 | 0.9238 | 0.5678 | 0.3931 | |
| H200002834 | FAM111A | 0.7931 | 1.1888 | 1.5798 | 2.7231 |
| H200008575 | MEF2A | 0.9956 | 1.0874 | 1.2269 | 2.1852 |
| H200001006 | DHRS13 | 0.8267 | 1.4287 | 1.8371 | 2.1821 |
| H200017445 | SUCNR1 | 0.8517 | 0.8042 | 1.2275 | 2.6318 |
| H200003843 | SAT1 | 0.9061 | 0.9701 | 1.1082 | 2.316 |
| H200004069 | PTPRE | 0.8568 | 0.9531 | 1.098 | 2.1504 |
| H200006587 | RGS2 | 0.6477 | 0.8868 | 1.4771 | 2.6675 |
| H200008209 | NAV3 | 0.6571 | 0.4408 | 1.253 | 3.3403 |
| H200007845 | 0.9676 | 0.4938 | 0.6507 | 1.2268 | |
| H200021128 | TCTN1 | 0.848 | 0.3728 | 0.4892 | 1.2922 |
| H200001569 | RSAD1 | 0.9789 | 0.9484 | 0.8524 | 0.4888 |
| H200017562 | C1GALT1C1 | 0.9565 | 0.7658 | 0.6933 | 0.4991 |
| H200002814 | C9orf140 | 0.9387 | 0.7971 | 0.7279 | 0.4626 |
| H200002245 | C22orf9 | 0.8398 | 0.8835 | 0.908 | 0.4672 |
| H200020390 | FAM46A | 0.8073 | 0.7061 | 0.6584 | 0.4897 |
| H200003471 | YWHAG | 0.852 | 0.52 | 0.4867 | 0.5355 |
| H200000419 | CENPA | 0.7722 | 0.542 | 0.4533 | 0.6846 |
| H200001941 | FAM46A | 0.7428 | 0.6849 | 0.6931 | 0.4513 |
| H200016105 | 0.7293 | 0.6981 | 0.4905 | 0.792 | |
| H200008360 | DHRS9 | 0.61 | 0.4867 | 0.6395 | 0.7368 |
| H200004769 | HIST1H4C | 0.5819 | 0.4728 | 0.6094 | 0.6927 |
| H200019346 | 0.6052 | 0.4432 | 0.4016 | 0.718 | |
| H200018740 | 0.5938 | 0.4321 | 0.5277 | 0.7306 | |
| H200001067 | EVI2B | 0.6304 | 0.4167 | 0.6691 | 0.7823 |
| H200016261 | STK6/AURKA | 0.6082 | 0.4028 | 0.441 | 0.6585 |
| H200013081 | DEPDC1 | 0.5525 | 0.3815 | 0.4393 | |
| H200020123 | 1.0639 | 1.2445 | 2.0239 | ||
| H200001749 | CASP7 | 0.9548 | 1.4103 | 2.544 | |
| H200007958 | CCDC134 | 0.694 | 0.7555 | 2.0645 | |
| H200018675 | 0.8948 | 0.8359 | 0.4542 | ||
| H200015636 | 2.0966 | 1.5265 | |||
| H200000156 | 5.1151 | 25.8726 | 126.9222 | ||
| H200001516 | PELI1 | 1.4342 | 1.7191 | 2.1665 | |
| H200011363 | 0.6085 | 0.4937 | |||
| H200000205 | HIST2H2AA3 | 0.4501 | 0.4727 | ||
| H200015535 | FICD | 2.263 | 2.2602 | ||
| H200016527 | CXorf34/TRMT2B | 2.0672 | 2.1392 | ||
| H200005739 | NEFM | 1.4999 | 2.8711 | ||
| H200001150 | ARRDC4 | 2.3531 | 1.9712 | ||
| H200016414 | 2.0232 | ||||
| H200017236 | LIMK2 | 1.4211 | 2.0302 | ||
| H200004185 | CDKN2AIP | 1.8434 | 2.2906 | ||
| H200003339 | AVPI1 | 1.8719 | 2.0933 | ||
| H200019676 | LRP11 | 2.005 | 1.9775 | ||
| H200004016 | PARP16 | 2.4133 | 2.2154 | ||
| H200000467 | SLC18A2 | 2.52 | 1.9316 | ||
| H200005857 | VLDLR | 3.4439 | 3.134 | ||
| H200004390 | DMC1 | 2.0052 | |||
| H200012875 | KIF1B | 2.5715 | |||
| H200008545 | TMEM9 | 2.192 | |||
| H200004439 | FAM89A | 2.3003 | |||
| H200016561 | PAG1 | 2.8081 | |||
| H200004662 | PELI2 | 2.0424 | |||
| H200012241 | TMEM154 | 2.2427 | |||
| H200009493 | CYB5R1 | 2.5233 | |||
| H200018222 | GPR84 | 2.3565 | |||
| H200001460 | SLC2A3 | 2.8812 | |||
| H200018921 | OSBPL6 | 2.0907 | |||
| H200006320 | SDS | 2.0108 | |||
| H200002343 | MASTL | 2.0644 | |||
| H200010023 | CPEB4 | 2.2967 | |||
| H200004544 | SERPINB8 | 2.8708 | |||
| H200000525 | PTX3 | 3.1971 | |||
Acknowledgments
This work was supported by the grant of National HighTech Program of China (No 2006AA020701). We are grateful to Dr Caiming Xu of Peking Union Medical College for providing the NB4 cell line. We thank Rui Ma and Chao Han for helping with the dielectrophoretic system setup. We also thank Dr Lan Xie and Dr Lei Wang for their helpful discussions and critical reading of the manuscript.
Footnotes
Disclosure
The authors have no conflicts of interest to disclose.
References
- 1.Grant S. Ara-C: Cellular and molecular pharmacology. Adv Cancer Res. 1998;72:197–233. doi: 10.1016/s0065-230x(08)60703-4. [DOI] [PubMed] [Google Scholar]
- 2.Schlenk RF, Germing U, Hartmann F, et al. High-dose cytarabine and mitoxantrone in consolidation therapy for acute promyelocytic leukemia. Leukemia. 2005;19(6):978–983. doi: 10.1038/sj.leu.2403766. [DOI] [PubMed] [Google Scholar]
- 3.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267(5203):1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 4.Ly JD, Grubb DR, Lawen A. The mitochondrial membrane potential (ΔΨm) in apoptosis; an update. Apoptosis. 2003;8(2):115–128. doi: 10.1023/a:1022945107762. [DOI] [PubMed] [Google Scholar]
- 5.Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol. 1992;148(7):2207–2216. [PubMed] [Google Scholar]
- 6.van Engeland M, Kuijpers HJH, Ramaekers FCS, Reutelingsperger CPM, Schutte B. Plasma membrane alterations and cytoskeletal changes in apoptosis. Exp Cell Res. 1997;235(2):421–430. doi: 10.1006/excr.1997.3738. [DOI] [PubMed] [Google Scholar]
- 7.Wyllie AH. Apoptosis (The 1992 Rose, Frank Memorial Lecture) Br J Cancer. 1993;67(2):205–208. doi: 10.1038/bjc.1993.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bortner CD, Cidlowski JA. Uncoupling cell shrinkage from apoptosis reveals that Na+ influx is required for volume loss during programmed cell death. J Biol Chem. 2003;278(40):39176–39184. doi: 10.1074/jbc.M303516200. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez M, Schaper J. Apoptosis: measurement and technical issues. J Mol Cell Cardiol. 2005;38(1):15–20. doi: 10.1016/j.yjmcc.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Salvioli S, Ardizzoni A, Franceschi C, Cossarizza A. JC-1, but not DiOC6(3) or rhodamine 123, is a reliable fluorescent probe to assess ΔΨm changes in intact cells: implications for studies on mitochondrial functionality during apoptosis. FEBS Lett. 1997;411(1):77–82. doi: 10.1016/s0014-5793(97)00669-8. [DOI] [PubMed] [Google Scholar]
- 11.Vermes I, Haanen C, Steffensnakken H, Reutelingsperger CJ. A novel assay for apoptosis flow cytometric detection of phosphatidylserine early apoptotic cells using fluorescein labeled expression on Annexin V. J Immunol Methods. 1995;184(1):39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- 12.Sanchis A, Brown AP, Sancho M, et al. Dielectric characterization of bacterial cells using dielectrophoresis. Bioelectromagnetics. 2007;28(5):393–401. doi: 10.1002/bem.20317. [DOI] [PubMed] [Google Scholar]
- 13.Gascoyne PRC, Noshari J, Anderson TJ, Becker FF. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis. 2009;30(8):1388–1398. doi: 10.1002/elps.200800373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang XB, Yang J, Huang Y, Vykoukal J, Becker FF, Gascoyne PRC. Cell separation by dielectrophoretic field-flow-fractionation. Anal Chem. 2000;72(4):832–839. doi: 10.1021/ac990922o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labeed FH, Coley HM, Thomas H, Hughes MP. Assessment of multidrug resistance reversal using dielectrophoresis and flow cytometry. Biophys J. 2003;85(3):2028–2034. doi: 10.1016/S0006-3495(03)74630-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoettges KF, Hübner Y, Broche LM, Ogin SL, Kass GEN, Hughes MP. Dielectrophoresis-activated multiwell plate for label-free high-throughput drug assessment. Anal Chem. 2008;80(6):2063–2068. doi: 10.1021/ac702083g. [DOI] [PubMed] [Google Scholar]
- 17.Valley JK, Swinton P, Boscardin WJ, et al. Preimplantation mouse embryo selection guided by light-induced dielectrophoresis. PLoS ONE. 2010;5(4):e10160. doi: 10.1371/journal.pone.0010160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flanagan LA, Lu J, Wang L, et al. Unique dielectric properties distinguish stem cells and their differentiated progeny. Stem Cells. 2008;26(3):656–665. doi: 10.1634/stemcells.2007-0810. [DOI] [PubMed] [Google Scholar]
- 19.Vykoukal J, Vykoukal DM, Freyberg S, Alt EU, Gascoyne PRC. Enrichment of putative stem cells from adipose tissue using dielectrophoretic field-flow fractionation. Lab Chip. 2008;8(8):1386–1393. doi: 10.1039/b717043b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chin S, Hughes MP, Coley HM, Labeed FH. Rapid assessment of early biophysical changes in K562 Cells during apoptosis determined using dielectrophoresis. Int J Nanomedicine. 2006;1(3):333–337. [PMC free article] [PubMed] [Google Scholar]
- 21.Labeed FH, Hughes MP, Thomas H, Coley HM. Rapid assay of apoptotic progress of mixed populations by dielectrophoresis; Proceedings of the American Association for Cancer Research Annual Meeting; Orlando, FL, USA. March 27–31, 2004. [Google Scholar]
- 22.Labeed FH, Coley HM, Hughes MP. Differences in the biophysical properties of membrane and cytoplasm of apoptotic cells revealed using dielectrophoresis. Biochim Biophys Acta. 2006;1760(6):922–929. doi: 10.1016/j.bbagen.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 23.Pethig R, Talary MS. Dielectrophoretic detection of membrane morphology changes in Jurkat T-cells undergoing etoposide-induced apoptosis. IET Nanobiotechnol. 2007;1(1):2–9. doi: 10.1049/iet-nbt:20060018. [DOI] [PubMed] [Google Scholar]
- 24.Wang XJ, Becker FF, Gascoyne PRC. Membrane dielectric changes indicate induced apoptosis in HL-60 cells more sensitively than surface phosphatidylserine expression or DNA fragmentation. Biochim Biophys Acta. 2002;1564(2):412–420. doi: 10.1016/s0005-2736(02)00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broche LM, Labeed FH, Hughes MP. Extraction of dielectric properties of multiple populations from dielectrophoretic collection spectrum data. Phys Med Biol. 2005;50(10):2267–2274. doi: 10.1088/0031-9155/50/10/006. [DOI] [PubMed] [Google Scholar]
- 26.Yu J, Zhang L, Chen AL, et al. Identification of the gene transcription and apoptosis mediated by TGF-beta-Smad2/3-Smad4 signaling. J Cell Physiol. 2005;215(2):422–433. doi: 10.1002/jcp.21325. [DOI] [PubMed] [Google Scholar]
- 27.Yang YH, Dudoit S, Luu P, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30(4):1–10. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29(4):365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 29.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei U, Sun PH, Pethig R. Refinement of the theory for extracting cell dielectric properties from dielectrophoresis and electrorotation experiments. Biomicrofluidics. 2011;5(4):044109. doi: 10.1063/1.3659282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lei U, Lo YJ. Review of the theory of generalised dielectrophoresis. IET Nanobiotechnol. 2011;5(3):86–106. doi: 10.1049/iet-nbt.2011.0001. [DOI] [PubMed] [Google Scholar]
- 32.Arnold WM, Klarmann BG, Sukhorukov VL, Zimmermann U. Membrane accommodation in hypo-osmotically-treated, and in giant electrofused cells. Biochem Soc Trans. 1992;20(2):120S. doi: 10.1042/bst020120s. [DOI] [PubMed] [Google Scholar]
- 33.Gascoyne PRC, Vykoukal JV. Dielectrophoresis-based sample handling in general-purpose programmable diagnostic instruments. Proc IEEE Inst Electr Electron Eng. 2004;92(1):22–42. doi: 10.1109/JPROC.2003.820535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sukhorukov VL, Arnold WM, Zimmermann U. Hypotonically induced changes in the plasma membrane of cultured mammalian cells. J Membr Biol. 1993;132(1):27–40. doi: 10.1007/BF00233049. [DOI] [PubMed] [Google Scholar]
- 35.Wang XB, Huang Y, Wang XJ, Becker FF, Gascoyne PRC. Dielectrophoretic manipulation of cells with spiral electrodes. Biophys J. 1997;72(4):1887–1899. doi: 10.1016/S0006-3495(97)78834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doherty GJ, McMahon HT. Mediation, modulation, and consequences of membrane-cytoskeleton interactions. Annu Rev Biophys. 2008;37:65–95. doi: 10.1146/annurev.biophys.37.032807.125912. [DOI] [PubMed] [Google Scholar]
- 37.Domnina LV, Ivanova OY, Cherniak BV, Skulachev VP, Vasiliev JM. Effects of the inhibitors of dynamics of cytoskeletal structures on the development of apoptosis induced by the tumor necrosis factor. Biochemistry (Mosc) 2002;67(7):737–746. doi: 10.1023/a:1016336421582. [DOI] [PubMed] [Google Scholar]
- 38.Flusberg DA, Numaguchi Y, Ingber DE. Cooperative control of Akt phosphorylation, bcl-2 expression, and apoptosis by cytoskeletal microflaments and microtubules in capillary endothelial cells. Mol Biol Cell. 2001;12(10):3087–3094. doi: 10.1091/mbc.12.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai F, Fernald AA, Zhao ND, Le Beau MM. cDNA cloning, expression pattern, genomic structure and chromosomal location of RAB6KIFL, a human kinesin-like gene. Gene. 2000;248(1–2):117–125. doi: 10.1016/s0378-1119(00)00135-9. [DOI] [PubMed] [Google Scholar]
- 40.Sardar HS, Luczak VG, Lopez MM, et al. Mitotic kinesin CENP-E promotes microtubule plus-end elongation. Curr Biol. 2010;20(18):1648–1653. doi: 10.1016/j.cub.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Putkey FR, Cramer T, Morphew MK, et al. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev Cell. 2002;3(3):351–365. doi: 10.1016/s1534-5807(02)00255-1. [DOI] [PubMed] [Google Scholar]
- 42.Hübner Y, Hoettges KF, Kass GEN, Ogin SL, Hughes MP. Parallel measurements of drug actions on Erythrocytes by dielectrophoresis, using a three-dimensional electrode design. IEE Proc Nanobiotechnol. 2005;152(4):150–154. doi: 10.1049/ip-nbt:20050011. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto K, Korenaga R, Kamiya A, Ando J. Fluid shear stress activates Ca2+ influx into human endothelial cells via P2X4 purinoceptors. Circ Res. 2000;87(5):385–391. doi: 10.1161/01.res.87.5.385. [DOI] [PubMed] [Google Scholar]
- 44.Zhang YJ, Sanchez D, Gorelik J, et al. Basolateral P2X4-like receptors regulate the extracellular ATP-stimulated epithelial Na+ channel activity in renal epithelia. Am J Physiol Renal Physiol. 2007;292(6):F1734–F1740. doi: 10.1152/ajprenal.00382.2006. [DOI] [PubMed] [Google Scholar]
- 45.Kobertz WR, Williams C, Miller C. Hanging gondola structure of the T1 domain in a voltage-gated K(+) channel. Biochemistry. 2000;39(34):10347–10352. doi: 10.1021/bi001292j. [DOI] [PubMed] [Google Scholar]
- 46.Miller C. An overview of the potassium channel family. Genome Biol. 2000;1(4):Reviews 0004.1–0004.5. doi: 10.1186/gb-2000-1-4-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bard AJ, Faulkner LR. Electrochemical Methods: Fundamentals and Applications. 2nd ed. New York, NY: Wiley; 2001. [Google Scholar]
- 48.Lodish H, Berk A, Zipursky LS, Matsudaira P, Baltimore D, Darnell J. Molecular Cell Biology. 4th ed. New York, NY: W.H.Freeman and Company; 2000. [Google Scholar]
- 49.Ashley RH. Challenging accepted ion channel biology: p64 and the CLIC family of putative intracellular anion channel proteins. Mol Membr Biol. 2003;20(1):1–11. doi: 10.1080/09687680210042746. [DOI] [PubMed] [Google Scholar]
- 50.Littler DR, Assaad NN, Harrop SJ, et al. Crystal structure of the soluble form of the redox-regulated chloride ion channel protein CLIC4. FEBS J. 2005;272(19):4996–5007. doi: 10.1111/j.1742-4658.2005.04909.x. [DOI] [PubMed] [Google Scholar]
- 51.Zittoun J, Marquet J, David JC, Maniey D, Zittoun R. A study of the mechanisms of cytotoxicity of Ara-C on three human leukemic cell lines. Cancer Chemother Pharmacol. 1989;24(4):251–255. doi: 10.1007/BF00257628. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Inhibition of growth of NB4 cells by Ara-C.
Note: Relative viabilities were determined using CCK-8 assays as described in the experimental section and the results were compared with untreated (control) cells.
Abbreviations: Ara-C, cytosine arabinoside; CCK-8, Cell Counting Kit-8.
Example of FCA of a single cell apoptosis experiment.
Note: Five time points are shown for FCA, 0, 2, 4, 6, and 12 hours after addition of 10−5 M Ara-C; PI+/FITC+ indicates deeply apoptotic or dead cells, while PI−/FITC+ indicates apoptotic cells.
Abbreviations: Ara-C, cytosine arabinoside; FCA, flow cytometric analysis; h, hours.
Gene expression profiling using microarray. (A) Gene ontology image of differentially expressed genes. (B) Cluster image of gene expression profiles to Ara-C-induced apoptosis from 0- to 12-hour time points.
Abbreviations: Ara-C, cytosine arabinoside; h, hours.
Real-time quantitative PCR analysis of the expression of selected genes. The results of real-time quantitative PCR analysis were similar to those microarray gene expression data. (A) Apoptosis-related genes. (B) Cell division- and proliferation-related genes. (C) Microtubule-related genes. (D) Ion transport-related genes.
Abbreviations: PCR, polymerase chain reaction.
Systematic transcription regulation pathway map of the apoptosis time course.
Notes: Solid dark lines represent previously known pathways; dashed dark lines represent undefined pathways. Red rectangles represent induced genes with fold changes ≥ 1.5; green rectangles represent repressed genes with fold changes ≥ 0.67. Blue ovals represent hypothetical activated TFs. Further TF ELISA experiments are needed to determine whether p53 or NF-κB activation is involved in Ara-C-induced apoptosis of NB4 cells. All elements are approximately arranged relative to the time axis.
Abbreviations: Ara-C, cytosine arabinoside; NF-κB, nuclear factor κB; TF, transcription factor.
Changes in intracellular calcium, sodium, and potassium over the 12-h time course measured using FCA. (A) An increase in Fluo 4-AM (Ca2+) fluorescence from control cells indicates an increase in intracellular calcium. (B) An increase in SBFI-AM (Na+) fluorescence from control cells indicates an increase in intracellular sodium. (C) A decrease in PBFI-AM (K+) fluorescence from control cells indicates a loss of intracellular potassium.
Notes: The data are shown as histograms which are representative of a single experiment.
Abbreviations: FCA, flow cytometric analysis h, hours.
Sequences of primer pairs used for real-time quantitative PCR
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| SESN2 | AGATGGAGAGCCGCTTTGAGCT | CCGAGTGAAGTCCTCATATCCG |
| E2F2 | AGCTGGAACCGAGAGAACATG | ACACGACCAGGCGAAACC |
| TNF | GGCGTGGAGCTGAGAGATAAC | GGTGTGGGTGAGGAGCACAT |
| HSPA1A | ACCTTCGACGTGTCCATCCTGA | TCCTCCACGAAGTGGTTCACCA |
| HSPA1B | ACCTTCGACGTGTCCATCCTGA | TCCTCCACGAAGTGGTTCACCA |
| C20orf129 | GCGTGGCAACAGGCTCCTACA | GGCATACAGGATTCGGAACTCC |
| IRS2 | CCTGCCCCCTGCCAACACCT | TGTGACATCCTGGTGATAAAGCC |
| DDIT4 | AGGAAGCTCATTGAGTTGTG | GGTACATGCTACACACACAT |
| KIF20A | GTACCAACCAGGAAAATCAG | TGTCTGAGTATTGCATCCTG |
| CENPE | GGAGAAAGATGACCTACAGAGGC | AGTTCCTCTTCAGTTTCCAGGTG |
| P2RX4 | GTGGCGGATTATGTGATACCAGC | CACACAGTGGTCGCATCTGGAA |
| KCTD9 | GCCGCTGTAATCTTGCACATGC | CAGTTTCAGGGATGCTCCTTCTG |
| GAPDH | CGCTCTCTGCTCCTCCTGTT | CCATGGTGTCTGAGCGATGT |
Abbreviation: PCR, polymerase chain reaction.
Number of differentially expressed genes
| Time (hours) | Up | Down | Total |
|---|---|---|---|
| 2 | 5 | 4 | 9 |
| 4 | 15 | 22 | 37 |
| 6 | 24 | 18 | 42 |
| 12 | 95 | 22 | 117 |
Total of 152 genes with ≥2.0-fold change at least once for all four time points
| Oligo_id | Gene | Ratio
|
|||
|---|---|---|---|---|---|
| 2 hours | 4 hours | 6 hours | 12 hours | ||
| H200015775 | TNF | 2.2768 | 1.6131 | 1.8894 | 2.1108 |
| H200019847 | DDIT3 | 1.1387 | 2.8434 | 3.7666 | 4.1329 |
| H200011743 | DDIT4 | 1.865 | 5.208 | 5.8026 | 5.2344 |
| H200001568 | SESN2 | 1.1782 | 2.5341 | 2.9756 | 2.1333 |
| H200008354 | TXNIP | 1.1029 | 2.4659 | 2.6146 | 3.029 |
| H200015433 | E2F2 | 1.3478 | 2.0973 | 2.3398 | 3.3365 |
| H200000027 | PMAIP1 | 1.2504 | 1.9617 | 3.1681 | 3.0868 |
| H200006759 | GADD45A | 1.11 | 1.7016 | 2.1676 | 2.1329 |
| H200012407 | CASP8AP2 | 1.4683 | 1.7928 | 2.1279 | 2.1613 |
| H200003690 | TRIB3 | 1.2974 | 1.9831 | 3.8015 | 3.8056 |
| H200006040 | CHAF1B | 1.2458 | 1.6792 | 2.2018 | 1.8877 |
| H200005977 | PRNP | 1.2409 | 1.2069 | 1.4649 | 2.3403 |
| H200006833 | NFKBIA | 1.0624 | 1.1148 | 1.3696 | 2.2215 |
| H200006782 | IRF1 | 1.2729 | 1.2868 | 1.6145 | 2.0059 |
| H200000124 | ATF3 | 1.1052 | 1.6494 | 1.559 | 2.7846 |
| H200011520 | NAB1 | 1.3486 | 1.4977 | 1.6588 | 2.0771 |
| H200010637 | E2F8 | 1.1142 | 1.5773 | 1.8893 | 2.0674 |
| H200004695 | REL | 1.521 | 1.9931 | 2.2944 | |
| H200015131 | TNFAIP3 | 1.7875 | 3.1083 | 10.82 | |
| H200019156 | EGR1 | 5.9961 | |||
| H200003705 | RASSF1 | 1.2268 | 2.3082 | ||
| H200003433 | MOAP1 | 2.2819 | |||
| H200006172 | ASNS | 2.1505 | |||
| H200012696 | ZNF624 | 3.6537 | |||
| H200015127 | PRKCE | 2.055 | |||
| H200008824 | KLF6 | 3.2992 | |||
| H200006354 | BTG1 | 0.8407 | 1.1771 | 2.1741 | 3.1005 |
| H200006664 | NFIL3 | 0.8679 | 1.2632 | 1.9841 | 3.1347 |
| H200005485 | PAK1 | 0.9977 | 1.1359 | 1.4839 | 2.0311 |
| H200008426 | DUSP6 | 0.6807 | 0.7826 | 1.0939 | 2.3816 |
| H200001719 | HSPA1A | 0.4533 | 0.8086 | 0.6038 | 0.4123 |
| H200017080 | HSPA1B | 0.4746 | 0.8206 | 0.5659 | 0.3964 |
| H200005758 | EIF4EBP1 | 0.5687 | 0.3518 | 0.3128 | 0.4273 |
| H200007392 | ARHGDIA | 0.9244 | 0.8883 | 0.7877 | 0.4844 |
| H200001819 | CCNE1 | 1.4023 | 2.2679 | 2.2377 | 1.8367 |
| H200006902 | TIEG/KLF10 | 1.3368 | 1.8159 | 1.7091 | 2.5303 |
| H200000106 | ADM | 1.0576 | 2.0229 | ||
| H200000570 | CXCL10 | 10.3587 | |||
| H200013414 | IRS2 | 0.4683 | 0.7933 | 1.121 | 1.0842 |
| H200005718 | C20orf129/FAM83D | 0.491 | 0.4594 | 0.4546 | 0.5191 |
| H200001767 | TPX2 | 0.6455 | 0.4921 | 0.5083 | 0.6906 |
| H200006436 | DLG7/DLGAP5 | 0.765 | 0.4849 | 0.5174 | 0.445 |
| H200010905 | BUB1 | 0.6302 | 0.448 | 0.4873 | 0.5465 |
| H200012276 | ASPM | 0.7851 | 0.4425 | 0.5062 | 0.6396 |
| H200003347 | CCNB1 | 0.5973 | 0.425 | 0.4063 | 0.4447 |
| H200005304 | KNTC2/NDC80 | 0.716 | 0.398 | 0.514 | 0.8752 |
| H200006423 | PLK1 | 0.6491 | 0.35 | 0.334 | |
| H200004906 | CDCA8 | 0.7698 | 0.563 | 0.4997 | 0.6453 |
| H200005764 | ZNF339/OVOL2 | 0.7392 | 0.6969 | 0.4721 | 0.6271 |
| H200006136 | CENPE | 0.6114 | 0.3547 | 0.4507 | 0.5201 |
| H200005853 | KIF20A | 0.5375 | 0.3259 | 0.31 | 0.411 |
| H200012386 | ECT2 | 0.4648 | |||
| H200005775 | KCTD9 | 1.7487 | 2.7477 | 4.1513 | |
| H200018987 | P2RX4 | 1.1764 | 1.7371 | 2.8761 | |
| H200003474 | CLIC4 | 1.181 | 1.1757 | 1.6314 | 2.1938 |
| H200005870 | CCL3 | 2.417 | 3.5841 | ||
| H200006175 | CCL4 | 3.4727 | 3.9945 | ||
| H200003915 | TNFAIP6 | 11.9615 | |||
| H200020490 | 2.1693 | 2.0409 | 1.4841 | 1.6363 | |
| H200006467 | HMGCS1 | 2.1238 | 1.5023 | 1.1664 | 1.0935 |
| H200005214 | INSIG1 | 2.8237 | 1.373 | 1.1014 | 1.0765 |
| H200004003 | CCNE2 | 1.6662 | 2.6628 | 3.8329 | 3.4481 |
| H200002187 | TIPARP | 1.1651 | 2.0555 | 2.1295 | 2.3472 |
| H200017137 | CCL3L1 | 1.3932 | 1.2863 | 2.057 | 5.6597 |
| H200006526 | ATP2B1 | 1.141 | 1.0957 | 1.6069 | 2.9833 |
| H200002418 | FOXO3A | 1.1284 | 1.1058 | 1.5603 | 2.667 |
| H200001281 | HECA | 1.0933 | 1.1625 | 1.6718 | 2.8152 |
| H200005834 | 1.105 | 1.177 | 1.2049 | 2.2256 | |
| H200004109 | RSN | 1.0981 | 1.2093 | 1.6478 | 2.2426 |
| H200002137 | KIAA1147 | 1.1165 | 1.2148 | 1.4234 | 2.3482 |
| H200002754 | N4BP2 | 1.3414 | 1.2147 | 1.5133 | 2.1562 |
| H200014202 | RGS16 | 1.0903 | 1.2847 | 1.6735 | 3.4525 |
| H200011487 | CCNB1IP1 | 1.1918 | 1.2945 | 1.5251 | 2.2965 |
| H200012358 | MAP1LC3B | 1.0403 | 1.3294 | 1.9677 | 2.3889 |
| H200009957 | 1.1546 | 1.4168 | 1.7351 | 2.4842 | |
| H200011669 | C6orf48 | 1.1391 | 1.5785 | 1.4732 | 2.7148 |
| H200006727 | MMD2 | 1.764 | 1.6883 | 1.8576 | 2.657 |
| H200006646 | CD83 | 1.4052 | 1.7151 | 1.8771 | 2.8336 |
| H200002948 | ATP2B1 | 1.1653 | 0.9668 | 1.2938 | 2.2311 |
| H200007999 | GBP7 | 1.1948 | 0.9544 | 1.0879 | 2.2038 |
| H200007904 | PTPRC | 1.1887 | 0.8226 | 0.9554 | 2.1509 |
| H200006209 | C1orf56/MLLT11 | 1.2761 | 0.93 | 0.9883 | 2.2463 |
| H200017557 | OPN3 | 1.3918 | 1.1323 | 0.7939 | 0.4675 |
| H200009444 | 1.0565 | 1.0179 | 0.9952 | 0.4823 | |
| H200006408 | MAT2A | 1.1349 | 1.0642 | 0.6771 | 0.4931 |
| H200000511 | TBXAS1 | 1.0995 | 0.9085 | 0.7469 | 0.4983 |
| H200006231 | KIAA0182 | 1.1452 | 0.7109 | 0.6425 | 0.4688 |
| H200012473 | PCF11 | 1.1002 | 0.8868 | 0.6982 | 0.4149 |
| H200006260 | 1.1805 | 0.9238 | 0.5678 | 0.3931 | |
| H200002834 | FAM111A | 0.7931 | 1.1888 | 1.5798 | 2.7231 |
| H200008575 | MEF2A | 0.9956 | 1.0874 | 1.2269 | 2.1852 |
| H200001006 | DHRS13 | 0.8267 | 1.4287 | 1.8371 | 2.1821 |
| H200017445 | SUCNR1 | 0.8517 | 0.8042 | 1.2275 | 2.6318 |
| H200003843 | SAT1 | 0.9061 | 0.9701 | 1.1082 | 2.316 |
| H200004069 | PTPRE | 0.8568 | 0.9531 | 1.098 | 2.1504 |
| H200006587 | RGS2 | 0.6477 | 0.8868 | 1.4771 | 2.6675 |
| H200008209 | NAV3 | 0.6571 | 0.4408 | 1.253 | 3.3403 |
| H200007845 | 0.9676 | 0.4938 | 0.6507 | 1.2268 | |
| H200021128 | TCTN1 | 0.848 | 0.3728 | 0.4892 | 1.2922 |
| H200001569 | RSAD1 | 0.9789 | 0.9484 | 0.8524 | 0.4888 |
| H200017562 | C1GALT1C1 | 0.9565 | 0.7658 | 0.6933 | 0.4991 |
| H200002814 | C9orf140 | 0.9387 | 0.7971 | 0.7279 | 0.4626 |
| H200002245 | C22orf9 | 0.8398 | 0.8835 | 0.908 | 0.4672 |
| H200020390 | FAM46A | 0.8073 | 0.7061 | 0.6584 | 0.4897 |
| H200003471 | YWHAG | 0.852 | 0.52 | 0.4867 | 0.5355 |
| H200000419 | CENPA | 0.7722 | 0.542 | 0.4533 | 0.6846 |
| H200001941 | FAM46A | 0.7428 | 0.6849 | 0.6931 | 0.4513 |
| H200016105 | 0.7293 | 0.6981 | 0.4905 | 0.792 | |
| H200008360 | DHRS9 | 0.61 | 0.4867 | 0.6395 | 0.7368 |
| H200004769 | HIST1H4C | 0.5819 | 0.4728 | 0.6094 | 0.6927 |
| H200019346 | 0.6052 | 0.4432 | 0.4016 | 0.718 | |
| H200018740 | 0.5938 | 0.4321 | 0.5277 | 0.7306 | |
| H200001067 | EVI2B | 0.6304 | 0.4167 | 0.6691 | 0.7823 |
| H200016261 | STK6/AURKA | 0.6082 | 0.4028 | 0.441 | 0.6585 |
| H200013081 | DEPDC1 | 0.5525 | 0.3815 | 0.4393 | |
| H200020123 | 1.0639 | 1.2445 | 2.0239 | ||
| H200001749 | CASP7 | 0.9548 | 1.4103 | 2.544 | |
| H200007958 | CCDC134 | 0.694 | 0.7555 | 2.0645 | |
| H200018675 | 0.8948 | 0.8359 | 0.4542 | ||
| H200015636 | 2.0966 | 1.5265 | |||
| H200000156 | 5.1151 | 25.8726 | 126.9222 | ||
| H200001516 | PELI1 | 1.4342 | 1.7191 | 2.1665 | |
| H200011363 | 0.6085 | 0.4937 | |||
| H200000205 | HIST2H2AA3 | 0.4501 | 0.4727 | ||
| H200015535 | FICD | 2.263 | 2.2602 | ||
| H200016527 | CXorf34/TRMT2B | 2.0672 | 2.1392 | ||
| H200005739 | NEFM | 1.4999 | 2.8711 | ||
| H200001150 | ARRDC4 | 2.3531 | 1.9712 | ||
| H200016414 | 2.0232 | ||||
| H200017236 | LIMK2 | 1.4211 | 2.0302 | ||
| H200004185 | CDKN2AIP | 1.8434 | 2.2906 | ||
| H200003339 | AVPI1 | 1.8719 | 2.0933 | ||
| H200019676 | LRP11 | 2.005 | 1.9775 | ||
| H200004016 | PARP16 | 2.4133 | 2.2154 | ||
| H200000467 | SLC18A2 | 2.52 | 1.9316 | ||
| H200005857 | VLDLR | 3.4439 | 3.134 | ||
| H200004390 | DMC1 | 2.0052 | |||
| H200012875 | KIF1B | 2.5715 | |||
| H200008545 | TMEM9 | 2.192 | |||
| H200004439 | FAM89A | 2.3003 | |||
| H200016561 | PAG1 | 2.8081 | |||
| H200004662 | PELI2 | 2.0424 | |||
| H200012241 | TMEM154 | 2.2427 | |||
| H200009493 | CYB5R1 | 2.5233 | |||
| H200018222 | GPR84 | 2.3565 | |||
| H200001460 | SLC2A3 | 2.8812 | |||
| H200018921 | OSBPL6 | 2.0907 | |||
| H200006320 | SDS | 2.0108 | |||
| H200002343 | MASTL | 2.0644 | |||
| H200010023 | CPEB4 | 2.2967 | |||
| H200004544 | SERPINB8 | 2.8708 | |||
| H200000525 | PTX3 | 3.1971 | |||