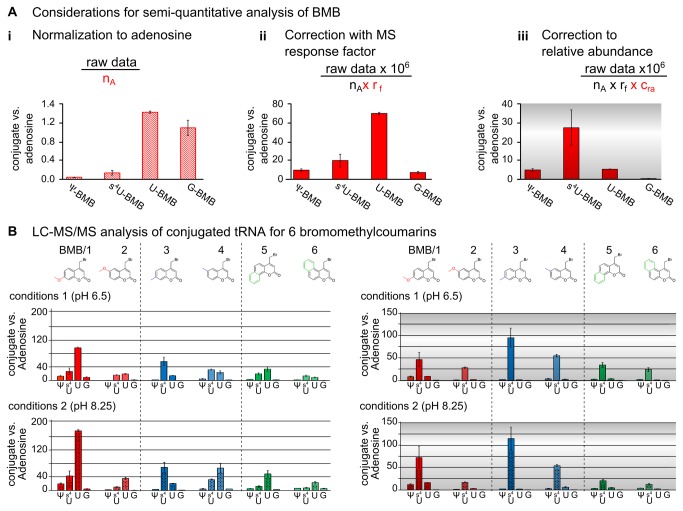
Figure 3. Data analysis of LC-MS/MS experiments of total tRNA E. coli treated with bromomethylcoumarins.
A) LC-MS/MS results from the chromatogram in Figure 2E after peak integration and data processing. The influence of the raw data processing steps is demonstrated for BMB conjugates. (i) Normalization to adenosine (nA) for intersample comparability reveals guanosine and uridine as the main reaction partners of BMB (ii). Usage of the found response factors (rf) which account for the differential ionization efficiencies in the mass spectrometer indicates U-BMB has the main reaction product. Data processing using nA and rf are used to display the reactivity of BMB (iii). Normalization to the relative abundance of tRNA modifications with the correction factor cra. This last data processing step is used to assess the selectivity of BMB for the substrate nucleosides. Here, 4-thiouridine is the main reaction partner with BMB.B) Reaction of BMB with total tRNA E. coli in comparison to 5 coumarins with different substitution patterns. On the top the chemical structure of all used coumarins are shown. The diagram below shows LC-MS/MS results of all coumarin-nucleoside conjugates from total tRNA E. coli reaction digests under slightly acidic conditions 1 (pH 6.5) monitored by their respective mass transitions (see Table S1-S6 in File S1) using the normalization to adenosine (factor nA). For comparison the mass integrals were corrected by usage of a response factor (rf) derived from absorption at 320 nm. The colors of the bars fit to the colors of the coumarin structures above. The graph displays the results for the reaction under more alkaline conditions 2 (pH 8.25), using the same data processing. Under these conditions the reactivity of guanosine with the coumarins is decreased and uridine is the prominent reaction partner. C) Reaction of all 6 differently substituted coumarins considering the nucleoside abundance (factor cra) by analysis of tRNA E. coli composition. The upper graph is for reaction conditions 1, the graph below for reaction conditions 2. The processed data clearly indicates a preference for 4-thiouridine of all tested coumarins which is most pronounced for reaction conditions 1 compared to conditions 2.