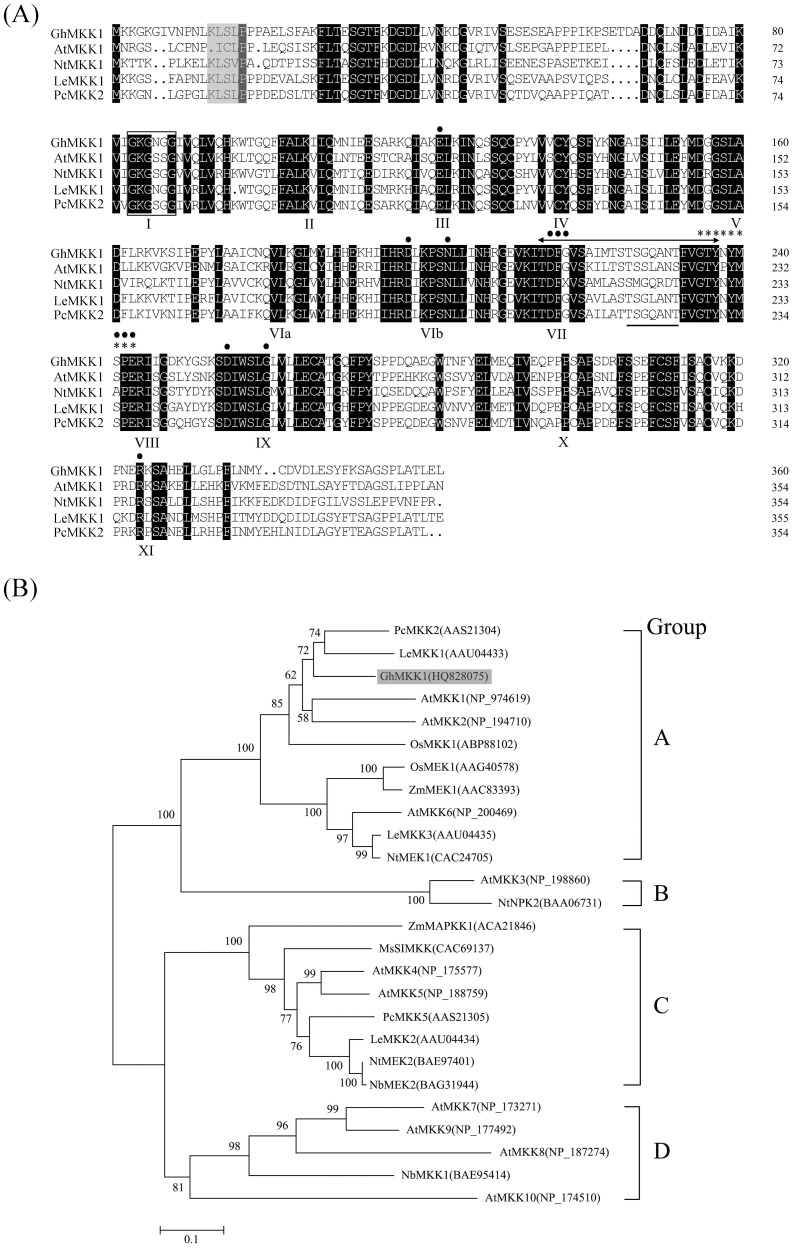
Figure 1. Characterization and sequence analysis of GhMKK1.
(A) Multiple amino acid sequence alignment of GhMKK1 (HQ828075) with AtMKK1 (NP_974619), NtMEK1 (CAC24705), LeMKK1 (AAU04433), and PcMKK2 (AAS21304). Identical and similar amino acids are shaded in black, and the docking domain is shaded in grey. The Roman numbers (I-XI) on the bottom indicate conserved subdomains, and a double-head arrow marks the active site motif. The serine and/or threonine residues in the conserved consensus motif, S/TXXXXXS/T, between subdomains VII and VIII of the MAPKKs are underlined, and the conserved consensus motif GXGXXG is boxed. The dots above the sequences represent activating sites, and asterisks indicate substrate specificity. (B) The phylogenetic relationships between GhMKK1 and other plant MAPKK proteins. The neighbor-joining phylogenetic tree was created using Clustal W and MEGA 4.0 software. The numbers above or below the branches indicate the bootstrap values (>50%) from 500 replicates. GhMKK1 is highlighted in grey. Each gene name is followed by its protein ID. The species of origin is indicated by the abbreviation before the gene names: Pc, Petroselinum crispum; Le, Lycopersicon esculentum; Gh, Gossypium hirsutum; At, Arabidopsis thaliana; Os, Oryza sativa; Zm, Zea mays; Nt, Nicotiana tabacum; Ms, Medicago sativa; and Nb, Nicotiana benthamiana. A, B, C, and D indicate the MAPKK group.