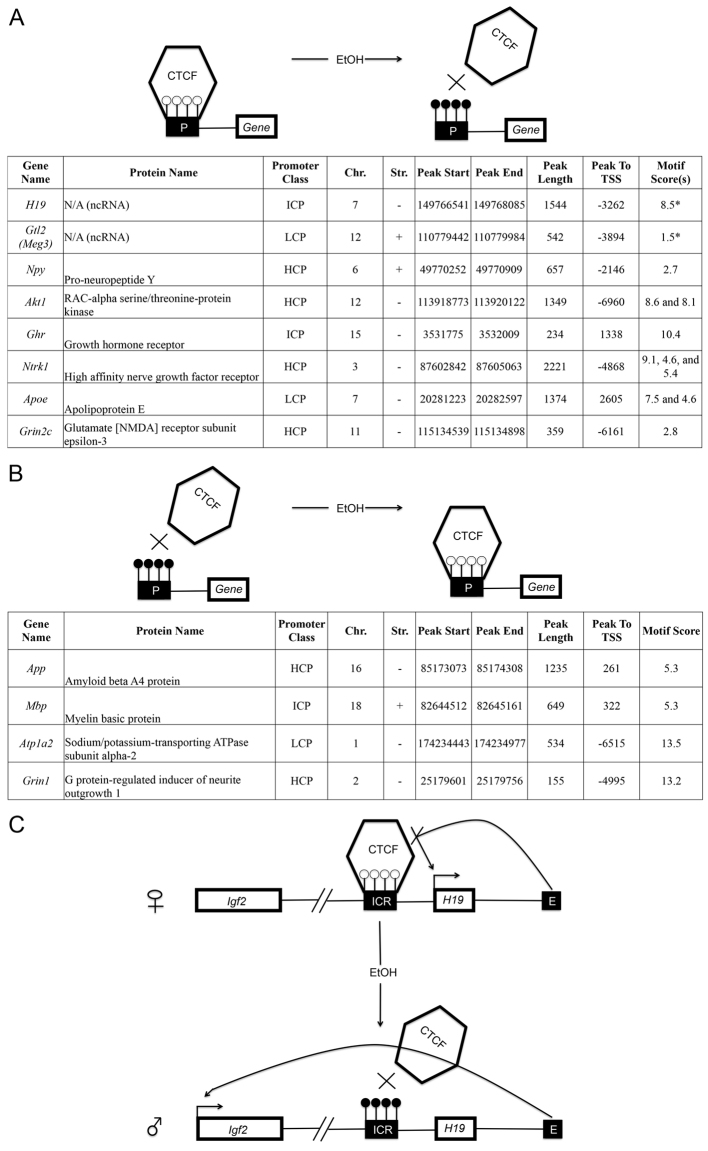
Fig. 2.
The functional significance of altered CTCF-binding-site methylation after FAE. (A,B) Gene promoters (‘P’) with CTCF sites showing increased (A) and decreased (B) methylation after FAE. (C) Schematic of H19/IGF2 imprinting regulation and the effects of FAE. The black rectangle represents the H19 ICR, white lollipops represent unmethylated DNA, and black lollipops represent methylated DNA. On the wild-type locus, the ICR exhibits paternal-specific methylation and contains binding sites for CTCF. On the maternal allele, CTCF binds to the ICR and blocks the Igf2 promoter from accessing the 3′ shared enhancers (E). On the paternal allele, the ICR is methylated, and H19 transcription is repressed. Because CTCF binding is methylation sensitive, the ICR cannot act as an insulator on the paternal allele, allowing Igf2 expression to be driven from the enhancer. Our results suggest that, in the case of FAE, imprinting is deregulated owing to increased methylation in the CTCF-binding site, which causes the maternal allele to exhibit paternal imprinting marks.