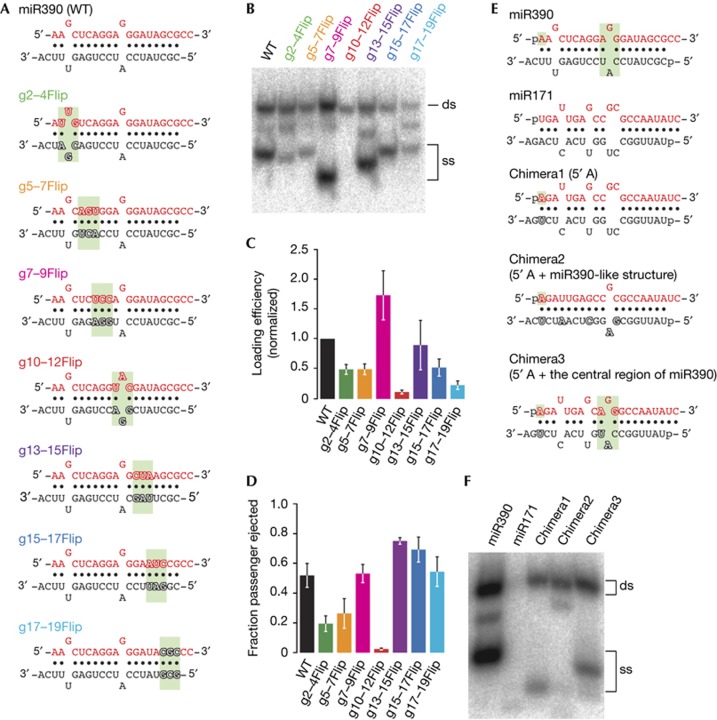
Figure 3.
The central region of miR390/miR390* is critical for AGO7–RISC assembly. (A) The structures of wild-type and flipped mutants of miR390/miR390* duplex. The flipped 3-nt regions are highlighted in green. The mutated nucleotides are outlined. (B–D) AGO7–RISC assembly using miR390 variants bearing flipped 3-nt sequences. The experiment was performed as in Fig 1A. (C,D) Quantification of duplex loading and passenger ejection efficiency in (B). The mean values±s.d. from three independent experiments are shown. (E) The structure of miR390/miR390*, miR171/miR171* and the chimeras between them. The 5′ A and the 3-nt central region of miR390/miR390* duplex are highlighted in green. The mutated nucleotides are outlined. (F) Introducing 5′ A to miR171/miR171* partially rescued duplex loading into AGO7, and the substitution of the central 3-nt region with that of miR390/miR390* produced mature AGO7–RISC. A, adenosine; AGO7, ARGONAUTE7; ds, double-stranded; nt, nucleotide; RISC, RNA-induced silencing complex; ss, single-stranded; WT, wild type.