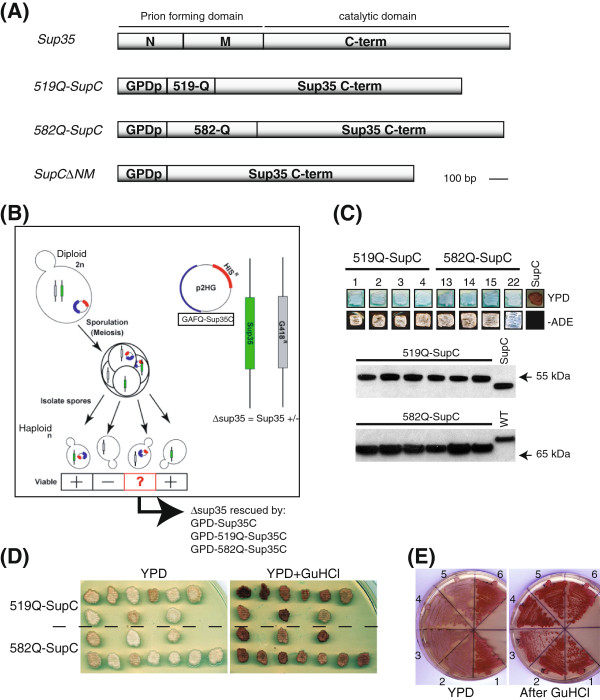
Figure 2.
Drosophila GAF polyglutamine domains (519Q and 582Q) substitute for prion domain of Sup35p in yeast. (A) Full length Sup35 regions encoding N, M and C term modules are indicated where N and M are known prion domain. GAF519 and GAF582 sequences encoding Q domains (519Q and 582Q) were fused to sequences encoding the C-terminal domain of Sup35p. The C-terminal of Sup35p alone (SupC) was used as a control. (B) Schematic picture depicting GAFQ-SupC chimeric (519Q-SupC and 582Q-SupC) constructs with HIS selectable marker tranfected in a diploid [PSI+] yeast strain, which was deleted for one copy of endogenous SUP35 indicated by G418 selectable marker. After induction of sporulation in transformants, the resulting haploids were selected on -His and G418 and analyzed for viability of individual spores. (C) The sup35∆ haploids with 519Q-SupC and 582Q-SupC fusions grow on YPD and -Ade media whereas SupC∆NM (SupC) containing haploid does not grow on -Ade and appear as red. The growth on -Ade plates indicates a nonsense suppression prion-like phenotype similar to [PSI+]. Expression levels of GAFQ-SupC fusions (519Q-SupC 57 kDa, 582Q-SupC 67 kDa, SupC∆NM 48 kDa) in individual haploids analyzed by Western blot with anti-Sup35 antibody recognizing only the C-terminal of Sup35. Total extract from wild type (WT) yeast shows endogenous Sup35 protein. (D) The prion-like phenotype of GAFQ-SupC expressing individual haploid cells is shown to be cured by 5 mM guanidine HCl (GuHCl), indicated by reversion of pink cells to red. (E) Streaking of GuHCl treated haploids show stability of non-sense suppression in two independent haploids of 519Q-SupC (2, 3) and 582Q-SupC (4, 5) taken from (D) which appear pink on YPD as compared to red colored haploids containing SupC∆NM (1,6). Both 519Q-SupC (2, 3) and 582Q-SupC (4, 5) haploids show curing after GuHCl and appear red as similar to SupC∆NM (1,6) haploids.