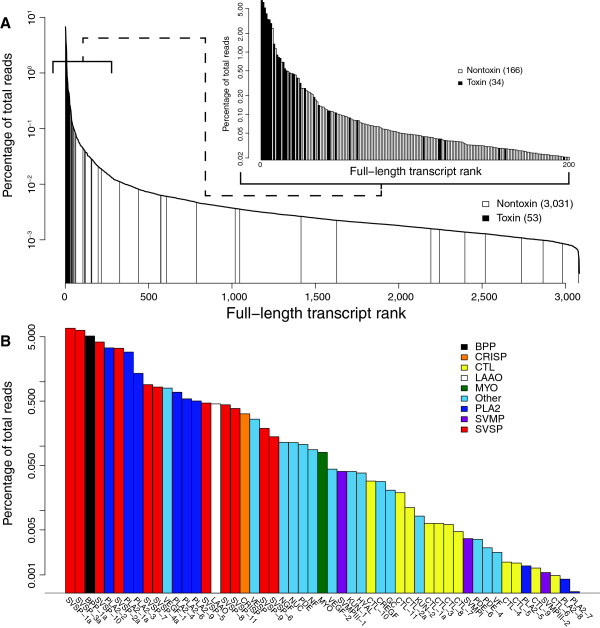
Figure 2.
The venom-gland transcriptome of Crotalus horridus was dominated by toxin transcripts. The 61 unique toxin transcripts were grouped into 53 clusters with less than 1% nucleotide divergence for this analysis. Note that all vertical axes are on log scales. (A) The overall expression patterns with all of the full-length annotated sequences showed that toxins dominated the high-abundance transcripts. The inset shows a magnification of the 200 most-abundant transcripts. (B) Expression levels of the toxin sequences showed a clear dominance of snake-venom serine proteinases (SVSPs), phospholipases A2 (PLA2s), and a single gene encoding bradykinin-potentiating and C-type natriuretic peptides (BPP). Expression levels among members of toxin classes were highly variable. The remaining toxin-class abbreviations are as follows: C-type lectin, CTL; cysteine-rich secretory protein, CRISP; cysteine-rich with EGF-like domain, CREGF; glutaminyl-peptide cyclotransferase, GC; hyaluronidase, HYAL; Kunitz-type protease inhibitor, KUN; L-amino acid oxidase, LAAO; myotoxin-A, MYO; nerve growth factor, NGF; neurotrophic factor, NF; nucleotidase, NUC; phosphodiesterase, PDE; snake-venom metalloproteinase, SVMP; vascular endothelial growth factor, VEGF; venom factor, VF; and vespryn, VESP.