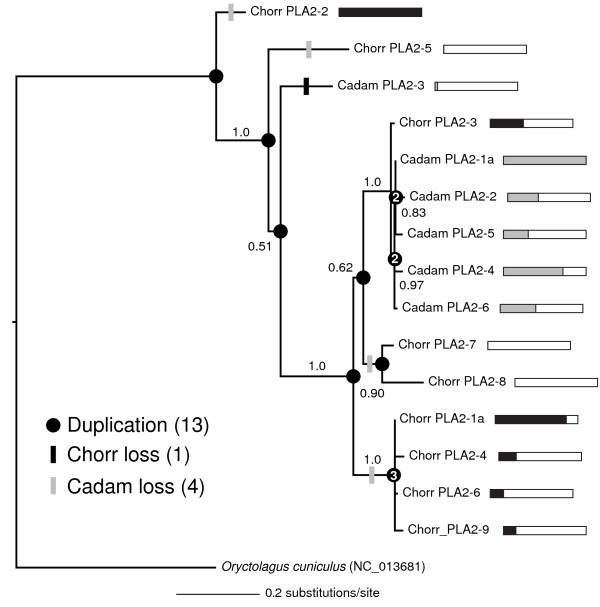
Figure 3.
Different clades were diversified and highly expressed in Crotalus adamanteus and C. horridus for the phospholipase A2 (PLA2) gene family. The bars adjacent to gene names give expression levels relative to the most highly expressed family member by species. A completely colored bar indicates the highest expression for the species. Clades comprising sequences from a single species either represent gene-family expansion for that species or gene-family contraction for the other. The four-gene clade including Chorr PLA2-1a and the five-gene clade including Cadam PLA2-1a are clear examples of this phenomenon. Duplication and gene-loss events were inferred by means of a duplication-loss parsimony model. We used a homologous nontoxin-PLA2 sequence from the european rabbit (Oryctolagus cuniculus) as an outgroup to root the phylogeny. Bayesian posterior probabilities are shown for clades for which the values exceeded 50%. Numbers within circles indicate that more than one duplication event was inferred for that node.