Figure 4. Identification of Alu motifs in promoters of ANRIL trans-regulated genes and ANRIL RNA and their spatial relation to PcG protein binding.
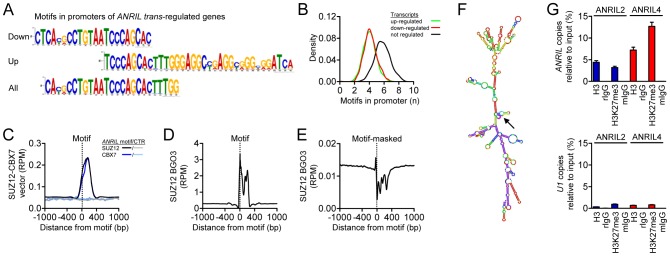
(A) DNA motif in promoters (5 kb) of trans-regulated genes representing an Alu-DEIN repeat [33]. (B) Number of Alu motifs in promoters of trans- and not regulated transcripts (n per 5 kb). (C) ChIP-seq enrichment of SUZ12 and CBX7 binding distal to Alu motif, demonstrating a specific spatial relation of motif occurrence to PcG protein binding. RPM- reads per million mapped reads, CTR- random DNA control sequence. (D) Motif-associated SUZ12 signal peaks in an independent data set from BGO3 cells. The actual signal peak only becomes apparent, if a multiple matching policy is adopted. (E) Using a strict unique-matches only policy, a substantial signal reduction is seen downstream of the motif. Please note differences in y-axis scaling in (D, E). (F) Secondary RNA structure prediction for ANRIL2 using the Vienna RNA package [58]. Within the minimum free energy structure, the Alu-DEIN motif is located in a stem-loop structure (arrow). (G) RIP demonstrating ANRIL binding to histone H3 (H3) and trimethylated lysine 27 of histone 3 (H3K27me3) in ANRIL2 (blue) and ANRIL4 (red) cells. U1 was used as negative control. mIgG/rIgG- mouse/rabbit IgG control. Error bars indicate s.e.m.
