Abstract
The endoplasmic reticulum (ER) functions to properly fold and process secreted and transmembrane proteins. Environmental and genetic factors that disrupt ER function cause an accumulation of misfolded and unfolded proteins in the ER lumen, a condition termed ER stress. ER stress activates a signaling network called the Unfolded Protein Response (UPR) to alleviate this stress and restore ER homeostasis, promoting cell survival and adaptation. However, under unresolvable ER stress conditions, the UPR promotes apoptosis. Here we discuss the current methods to measure ER stress levels, UPR activation, and subsequent pathways in mammalian cells. These methods will assist us in understanding the UPR and its contribution to ER stress related-disorders such as diabetes and neurodegeneration.
1. Introduction
The endoplasmic reticulum (ER) is a multifunctional organelle essential for the synthesis, folding, and processing of secretory and transmembrane proteins. In order for proteins to fold properly a balance between the ER protein load and the folding capacity to process this load must be established. However, physiological and pathological stimuli can disrupt this ER homeostasis resulting to an accumulation of misfolded and unfolded proteins, a condition known as ER stress. ER stress activates a complex signaling network referred as the Unfolded Protein response (UPR) to reduce ER stress and restore homeostasis. However, if the UPR fails to reestablish the ER to normality, ER stress causes cell dysfunction and death. [1]. Recent evidence further indicates that ER stress-mediated cell dysfunction and death is involved in pathogenesis of human chronic disorders including diabetes and neurodegeneraiton [2]. This chapter discusses the methods for measuring and quantifying ER stress levels, UPR activation and the subsequent downstream outcomes. We will mainly focus on the tissue culture system. Studying ER stress and the UPR will help us understand the pathophysiology and develop novel therapeutic modalities for ER stress-related disorders.
2. ER stress and the UPR
2.1. Endoplasmic reticulum (ER) and ER stress
The endoplasmic reticulum (ER) has an important role in the folding and maturation of newly synthesized secretory and transmembrane proteins. To ensure proper protein folding, the ER lumen maintains a unique environment to establish a balance between the ER protein load and the capacity to handle this load. This ER homeostasis can be perturbed by physiological and pathological insults such as high protein demand, viral infections, environmental toxins, inflammatory cytokines, and mutant protein expression resulting to an accumulation of misfolded and unfolded proteins in the ER lumen, a condition termed as ER stress.
2.2. The Unfolded Protein Response (UPR)
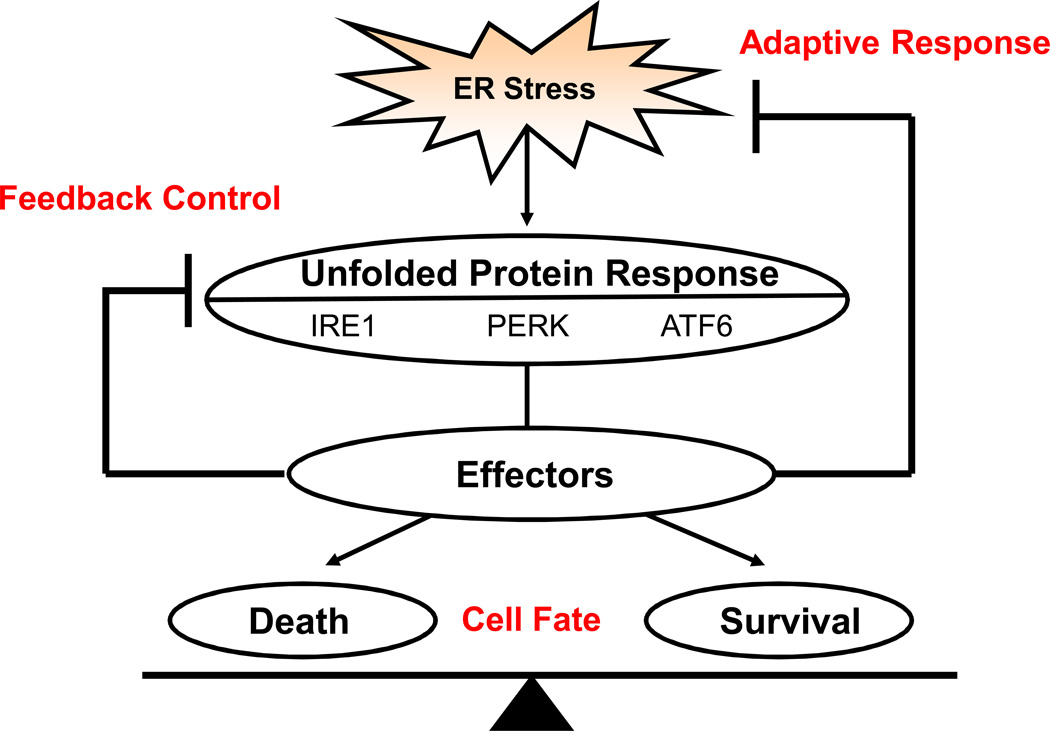
The adaptive response to ER stress is the unfolded protein response (UPR) (Figure 1). The UPR is initiated by three ER transmembrane proteins: Inositol Requiring 1 (IRE1), PKR-like ER kinase (PERK), and Activating Transcription Factor 6 (ATF6). During unstressed conditions, the ER chaperone, immunoglobin binding protein (BiP) binds to the luminal domains of these master regulators keeping them inactive. Upon ER stress, BiP dissociates from these sensors resulting to their activation.
Figure 1.
Response categories of the Unfolded Protein Response. Three ER transmembrane proteins, IRE1, PERK, and ATF6, sense ER stress in the ER lumen and become activated regulating a cascade of signaling pathways collectively termed the Unfolded Protein Response (UPR). The UPR has three functions: adaptive response, feedback control, and cell fate. Under the adaptive response, the UPR aims to reduce ER stress and restore ER homeostasis. If the UPR is successful, the UPR signaling pathways are turned off by feedback mechanisms. The UPR also regulates both survival and death factors that govern whether the cell will live or not depending on the severity of the ER stress condition.
The activated UPR regulates downstream effectors with the following three distinct functions: adaptive response, feedback control, and cell fate regulation [3]. (Figure 1) The UPR adaptive response includes upregulation of molecular chaperones and protein processing enzymes to increase folding and handling efficiency, translational attenuation to reduce ER workload and prevent further accumulation of unfolded proteins, and an increase in ER-associated protein degradation (ERAD) and autophagy components to promote clearance of unwanted proteins. Feedback control involves the negative regulation of UPR activation as ER homeostasis is being re-established to prevent harmful hyperactivation. . Cell fate regulation by the UPR plays an important role in the pathogenesis of ER stress-related disorder. Our current model is that the UPR directly regulates both apoptotic and anti-apoptotic outputs, acting as a binary switch between the life and death of ER stressed cells [3]. When the cell encounters ER stress that the UPR can mitigate, the cell will survive and is primed for future ER stress insults. However, during unresolvable ER stress conditions, the UPR fails to reduce ER stress and restore homeostasis promoting cell death.
2.3. IRE1, PERK, and ATF6 signaling pathways
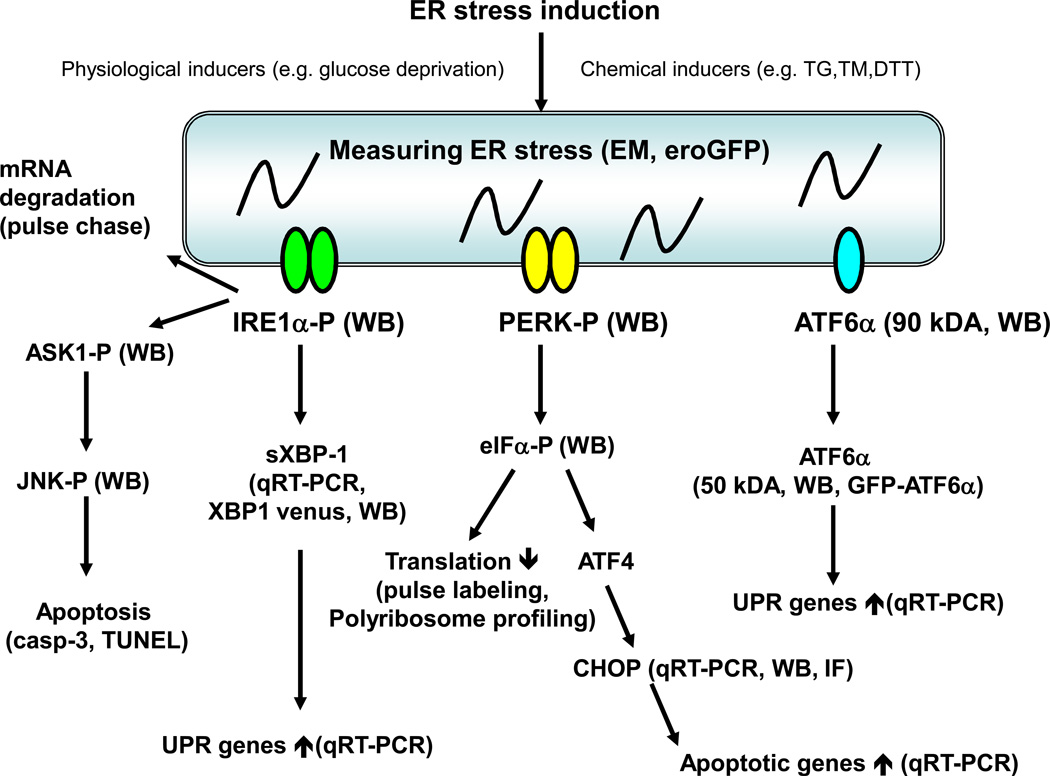
As mentioned previously, the UPR is regulated by the three master regulators, IRE1, PERK, and ATF6. (Figure 2)
Figure 2. Studying ER stress and the Unfolded Protein Response in Mammalian cells.
ER stress can be induced by several chemical and physiological inducers. Actual ER stress has been difficult to measure directly. Currently observing ER distension by electron microscopy (EM) and measuring oxidative protein folding (eroGFP) are available. The activation of the UPR master regulators has also been challenging but could be attempted by detecting IRE1α and PERK phosphorylation, and ATF6α cleavage by specific antibodies using western blot (WB). ATF6α translocation can be monitored by fluorescence microscopy of GFP-ATF6α. The downstream outputs of the UPR master regulars are readily measurable. IRE1α splices XBP-1 mRNA which can be detected by quantitative real time PCR (qRT-PCR), XBP-1 venus, and western blot (WB). IRE1α also mediates degradation of ER localized mRNAs, which can be measured by pulse chase assays. PERK phosphorylates eIF2α, which can be detected by specific antibodies. Phosphorylated eIF2α triggers global mRNA translation attenuation, which can be measured by standard methods such as pulse labeling and polyribsome profiling. Activated ATF6α is a transcription factor and regulation of its downstream target genes can be measured by qRT-PCR. The UPR in general regulates several transcription factors and in turn their transcriptional targets can be measured by qRT-PCR and luciferase assays. The UPR also induces expression of ERAD components as well as survival and death components. ERAD and apoptosis can be measured by standard methods.
IRE1, a type I ER transmembrane kinase, senses ER stress by its N-terminal luminal domain[4]. Upon sensing the presence of unfolded or misfolded proteins, IRE1 dimerizes and autophosphorylates to become active. There are two isoforms of IRE1: IRE1α and IRE1β. IRE1α is expressed in all cell types and has been extensively studied. Activated IRE1α splices X-box binding protein 1 (XBP1) mRNA [5–7]. Spliced XBP1 mRNA encodes a basic leucine zipper (b-ZIP) transcription factor that upregulates UPR target genes, including genes that function in ERAD such as ER-degradation-enhancing-α-mannidose-like protein (EDEM) [8], as well as genes that function in folding proteins such as protein disulfide isomerase (PDI) [9]. High levels of chronic ER stress can lead to the recruitment of TNF-receptor-associated factor 2 (TRAF2) by IRE1 and the activation of apoptosis-signaling-kinase 1 (ASK1). Activated ASK1 activates c-Jun N-terminal protein kinase (JNK), which in turn plays a role in apoptosis by regulating the BCL2 family of proteins [10–12].
PERK is also a type I ER transmembrane kinase. Similar to IRE1α, when activated by ER stress, PERK oligomerizes, autophosphorylates and then directly phosphorylates Ser51 on the α subunit of eukaryotic initiation factor 2 (eIF2α) [13]. Phosphorylated eIF2α prevents formation of ribosomal initiation complexes leading to global mRNA translational attenuation. This reduction in ER workload protects cells from ER stress-mediated apoptosis [14]. Meanwhile some mRNAs require eIF2α phosphorylation for translation such as the mRNA encoding activating transcription factor 4 (ATF4). ATF4 is a b-ZIP transcription factor that regulates several UPR target genes including those involved in ER stress-mediated apoptosis such as C/EBP homologous protein (CHOP) [15].
A third regulator of ER stress signaling is the type II ER transmembrane transcription factor, ATF6. [16]. ATF6 has two isoforms, ATF6α and ATF6β. ATF6α has been extensively studied in the context of ER Stress. Upon ER stress conditions, ATF6α transits to the Golgi where it is cleaved by site 1 (S1) and site 2 (S2) proteases, generating an activated b-ZIP factor [17]. This processed form of ATF6α translocates to the nucleus to activate UPR genes involved in protein folding, processing, and degradation. [18, 19].
3. Cell culture system and ER stress induction
3.1. Mammalian cells as a model system for studying ER stress and the UPR
Protein folding in the ER is essential to the survival of individual cells, explaining the evolution of the UPR in unicellular organisms such as yeast. But as secretion is the basis of multicellularity, ER protein folding homeostasis powerfully impacts the physiology of mammals. Dysregulation of ER homeostasis can cause chronic diseases in humans. Therefore, it is important to study ER stress and the UPR using mammalian cells to understand the UPR and ER stress-related diseases.
3.2. Examples of cell lines and primary cells that are used as a model
3.2.1. Mammalian cells
There are several mammalian cells lines that demonstrate a response to commonly applied ER stress inducers and the subsequent UPR activation. Mouse embryonic fibroblasts (MEFs) are powerful tools for studying the UPR because MEFs from Ire1α, Ire1β, Perk, ATf6α, ATF6β, Xbp-1, Atf4, and Gadd34 knockout mice are available [10, 20–27]. The UPR is particularly important for maintaining ER homeostasis in professional secretory cells such as pancreatic β cells and plasma cells. The mouse β cell line, MIN6, and the rat β cell lines, INS-1 and INS-1 832/13, are often used to study ER stress and the UPR in the context of the β cell [28–30]. Human, mouse, and rat primary islets are also great tools available. Multiple myeloma is a cancer derived from plasma cells. J558 is a multiple myloma cell line and has been shown to be useful to develop a therapy targeting the XBP-1 pathways [31]. It is important to note that each cell type responds to ER stress and activates the UPR in a unique manner.
3.3 ER stress inducers
3.3.1. Pharmaceutical ER stress inducers
There are several available chemicals to induce ER stress and activate the UPR in a tissue culture system including tunicamycin, thapsigargin, Brefedin A, dithiothreitol (DTT), and MG132. The concentration and duration of treatment in which ER stress is induced by these compounds should be determined for each particular system. Typically only a few hours are required to induce ER stress and long exposures often induce ER stress-mediated cell death. Tunicamycin is an inhibitor of the UDP-N-acetylglucosamine-dolichol phosphate N-acetylglucosamine-1-phosphate transferase (GPT), therefore blocking the initial step of glycoprotein biosynthesis in the ER. Thus, treatment of tunicamycin causes accumulation of unfolded glycoproteins in the ER, leading to ER stress. In many cell types, ER stress can be induced by treating cells with 2.5–5 µg/ml of tunicamycin for 5 hours. Thapsigargin is a specific inhibitor of the sarcoplasmic/endoplasmic reticulum Ca2+-ATPase (SERCA). Treatment with thapsigargin results in a decrease in ER calcium levels. When calcium levels are lowered in the ER, the calcium-dependent ER chaperones, such as calnexin, lose their chaperone activity, leading to the accumulation of unfolded proteins. ER stress can be induced by treating cells with 0.1–1 µM of thapsigargin for 5 hours. Brefeldin A inhibits transport of proteins from the ER to the Golgi and induces retrograde protein transport from the Golgi apparatus to the endoplasmic reticulum. This leads to the accumulation of unfolded proteins in the ER. DTT is a strong reducing agent and blocks disulfide-bond formation, quickly leading to ER stress within minutes. Because DTT also blocks disulfide-bond formation of newly synthesized proteins in the cytosol, it is not a specific ER stress inducer. MG132, is a specific, and cell-permeable proteasome inhibitor. Consequently MG132 blocks ER-associated protein degradation (ERAD) and causes misfolded proteins to accumulate in the ER. Thus, MG132 induces ER stress indirectly.
3.3.2 Physiological ER stress inducers
Physiological perturbants are commonly used to induce mild ER stress in a tissue culture system. As mentioned with pharmaceutical inducers, the amount and time of exposure to these inducers should be determined in a given system. Glucose deprivation blocks N-linked glycosylation and reduces cellular ATP levels, leading to ER stress in many cell types. ER stress can be induced by treating cells with glucose-free media for 24–48 hours. Glucose deprivation is not a strong inducer of cell death.
ER stress and the UPR are unique from cell to cell. Therefore there are specific ER stress inducers for a given cell type each activating the UPR in a distinct manner. ER stress and the UPR have been extensively studied in pancreatic β cells. Some specific β cell ER stress inducers include acute and chronic high glucose, cytokines, free fatty acids, and overexpression of mutant insulin-2. Both acute (1–3 hours) and chronic (≥ 24 hours) high glucose (≥16.7 mM) induces IRE1α phosphorylation in pancreatic β cells. However, acute high glucose does not induce PERK activation. Inflammatory cytokines such as IL-1β and IFN-γ can cause ER stress in β cell lines and primary islets. In rat insulinoma INS-1E cells and rat primary islets, ER stress can be triggered by IL-1β (50 units/ml) alone or IL-1β (50 units/ml) + IFN-γ (0.036 µg/ml) in 24 hours [32] activating the IRE1 and PERK arms of the UPR. Saturated free fatty acid, palmitate (0.25 – 5mM) can cause ER stress and activate the UPR within at least 4 hours in INS-1E rat insulinoma cells and MIN6 mouse insulinoma cells as well as in primary rat and human islets [33]. Finally β cells specialize in the synthesis, processing and secretion of insulin. Thus overexpression of misfolded mutant insulin-2 (C96Y) in β cell lines and pancreatic islets from Akita mice expressing this mutant insulin demonstrate ER stress and activation of the UPR. [34]
It has been shown that free cholesterol causes ER stress in macrophages and preferentially activates the CHOP branch of the UPR. Acetyl-LDL (100 µg/ml) plus the acyl-CoA:cholesterol acyltransferase inhibitor 58035 induces ER stress in macrophages in 5–10 hours [35]. This method is used to study the role of ER stress-mediated macrophage death during the progression of atherosclerosis [35].
In vascular endothelial cells, homocysteine can cause ER stress. In Human Umbilical Vein Endothelial Cells (HUVEC), GRP78 (BiP) mRNA expression is induced by 1–5 mM of homocysteine in 4–8 hours [36].
4. Measuring ER stress
Commonly we add an ER stress inducer and measure the activation of the UPR and the consequent downstream responses. However these results do not directly reflect the accumulation of misfolded and unfolded protein within the ER lumen. It has been challenging to directly measure ER stress levels in cells. Here we discuss at least two methods that directly measure ER stress.
4.1. ER dilation (EM)
Upon ER stress, the ER lumen is remarkably enlarged in cells and tissues, which can be detected by electron microscopy [37–39]. This method has been used often to detect ER stress in pancreatic β cells.
4.2. Real-time redox measurements during ER stress
The ER maintains an oxidizing environment to promote disulfide bond formation in newly synthesized proteins [40]. An increase in ER protein load could overwhelm oxidative folding enzymes, preventing proper disulfide formation and therefore inducing ER stress. Feroz Papa’s group recently developed a method to monitor the redox state of GFP to reflect ER stress [42]. This reporter named “eroGFP (ER-targeted redox-sensitive GFP)” has been designed to change fluorescence at two maximas, 400 and 490 nm, upon disulfide formation between an engineered cysteine pair. As eroGFP becomes reduced, excitation at 490 nm increases while decreases at 400 nm. The ratio of the fluorescence measured at 490 nm versus 400 nm reports ER redox status in cells.. Currently this method is only available in yeast cells. We are collaborating with Papa’s group to adapt this system in mammalian cells (Ishigaki, Marksamer, Lu, Papa, and Urano, unpublished).
5. Studying the UPR master regulators activation
The ability to measure IRE1α and PERK phosphorylation, and ATF6α cleavage would be ideal to determine UPR activation levels. However; endogenous expression levels of these molecules are low and hard to detect with available commercial antibodies. Thus, alternatively, we suggest measuring expression and activation levels of downstream components regulated by these master regulators to determine UPR activation.
5.1. Methods for measuring IRE1α activation
IRE1α is an ER transmembrane serine/threonine kinse undergoing autophosphoryaltion upon ER stress. Thus, the best way to measure activation levels of IRE1α is to measure its phosophorylation levels. Our group developed anti-phospho-IRE1α specific antibody from bulk antiserum by affinity purification followed by adsorption against the nonphospho analog column peptide (Open biosystems, Huntsville, AL). The peptide sequence for generating the antibody was CVGRH (pS) FSRRSG. This phosphopeptide was synthesized, multi-link-conjugated to KLH, and used to immunize 2SPF rabbits. Rabbit anti-total-IRE1α antibody (B9134) was generated using a peptide, EGWIAPEMLSEDCK. Samples are prepared by lysing cells with ice cold M-PER buffer (PIERCE, Rockford, IL) containing protease inhibitors, incubated on ice for 15 min and centrifuged at 13,000 × g for 15 min at 4°C. Supernatant was collected and total protein concentrations are measured. 10 ug of proteins are prepared with sample buffer and heated for 5 min at 95°C. Denatured proteins were separated using 4%–20% linear gradient SDS-PAGE (Bio Rad, Hercules, CA) and transferred onto PVDF membrane. Non-specific binding sites are blocked with 5% milk in 1X TBS + 0.1% Tween-20 (TBST) for 1 h at room temperature followed by overnight incubation at 4°C with diluted anti-IRE1α antibody (1:1000) in 5% milk-TBST. Next day, blots are incubated with diluted HRP-conjugated anti-rabbit antibody (1:3000, Cell Signaling) in 5% BSA-TBST for 1 h. Membrane–bound antibodies are detected by ECL Western Blotting Substrate (Pierce). All of our immunoblots are performed in this manner. Table 1 lists working antibodies that we commonly use to detect UPR proteins.
Table 1.
List of antibodies for detecting ER stress markers
| Antibody | Source | Weight (kDA) | Supplier | Blocking buffer, Dilution |
|---|---|---|---|---|
| IRE1α | Rabbit | 130 | Cell Signaling, #3294 | BSA, WB 1:1000 |
| Phospho-IRE1α | Rabbit | 110 | Novus, NB100-2323 | Milk, WB 1:1000 |
| Spliced XBP-1 | Rabbit | 54 | Santa Cruz, sc-7160 | Milk, WB 1:1000 |
| Total PERK | Rabbit | 150 | Rocklan, 100-401-962 | Milk, WB 1:1000 |
| Phospho-PERK | Rabbit | 170 | Cell Signaling, #3179 | BSA, WB 1:1000 |
| eIF2α | Rabbit | 36 | Santa Cruz, sc-11386 | Milk, WB 1:1000 |
| Phospho-eIF2α | Rabbit | 38 | Cell Signaling, #3597 | BSA, WB 1:1000 |
| ATF6α | Rabbit | 90 | Santa Cruz, sc-22799 | Milk. WB 1:100 |
| Cleaved ATF6α | Mouse | 50 | Imgenex, IMG-273 | Milk, WB 1:1000 |
| CHOP/GADD153 | Mouse | 31 | Pierce, MA1-250 | Milk, WB 1:2000 |
| BiP/GRP78 | Rabbit | 78 | Stressgen, SPA-826 | Milk, WB 1:1000 IF: 1:100 |
| ATF4 | Rabbit | 39 | ProteinTech, 10835-1-AP | Milk, WB 1:1000 |
| PDI | Mouse | 58 | Stressgen, SPA-891 | Milk, WB 1:1000 IF: 1:100 |
Activated IRE1α functions as an endoribonuclease splicing a 26 base pair intron from XBP-1 mRNA. Spliced XBP-1 mRNA is translated into a stable and active UPR transcription factor. Measuring XBP-1 splicing represents a reliable indirect method of determining IRE1α activation. We have designed primers that can specifically detect spliced and unspliced XBP-1 transcripts by quantitative real-time PCR. [CO2][43]. (Table 2) Using these primers, we could successfully quantify expression levels of spliced and unspliced XBP-1 in different cell lines [44, 45]. Total RNA were isolated from cells using RNeasy Mini Kit (Qiagen) and reverse transcribed to cDNA using 1 µg of total RNA with ImProm-II Reverse transcription system (Promega). Primers and diluted cDNA samples were prepared with Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) for qRT-PCR. For the thermal cycle reaction, the iQ5 system (BioRad, Hercules, CA) was used at 95°C for 10 min, then 40 cycles at 95°C for 10 sec and at 55°C for 30 sec. The relative amount for each transcript was calculated by a standard curve of cycle thresholds for serial dilutions of cDNA samples and normalized to the amount of β-actin. The polymerase chain reaction (PCR) was performed in triplicate for each sample, after which all experiments were repeated twice. Spliced and unspliced XBP-1 mRNA can also be measured by semi-quantitative RT-PCR and XBP-1 protein can be detected by immunoblot using anti-XBP-1 specific antibody (Santa Cruz).
Table 2.
List of primers for detecting ER stress markers by real-time PCR
| Gene | Human | Mouse | Rat |
|---|---|---|---|
| sXBP1 | CTGAGTCCGAATCAGGTGCAG ATCCATGGGGAGATGTTCTGG |
CTGAGTCCGAATCAGGTGCAG GTCCATGGGAAGATGTTCTGG |
CTGAGTCCGAATCAGGTGCAG ATCCATGGGAAGATGTTCTGG |
| usXBP1 | CAGCACTCAGACTACGTGCA ATCCATGGGGAGATGTTCTGG |
CAGCACTCAGACTATGTGCA GTCCATGGGAAGATGTTCTGG |
CAGCACTCAGACTACGTGCG ATCCATGGGAAGATGTTCTGG |
| Total XBP1 | TGGCCGGGTCTGCTGAGTCCG ATCCATGGGGAGATGTTCTGG |
TGGCCGGGTCTGCTGAGTCCG GTCCATGGGAAGATGTTCTGG |
TGGCCGGGTCTGCTGAGTCCG ATCCATGGGAAGATGTTCTGG |
| ATF4 | GTTCTCCAGCGACAAGGCTA ATCCTGCTTGCTGTTGTTGG |
GGGTTCTGTCTTCCACTCCA AAGCAGCAGAGTCAGGCTTTC |
AATGGATGACCTGGAAACCA TCTTGGACTAGAGGGGCAAA |
| CHOP | AGAACCAGGAAACGGAAACAGA TCTCCTTCATGCGCTGCTTT |
CCACCACACCTGAAAGCAGAA AGGTGAAAGGCAGGGACTCA |
AGAGTGGTCAGTGCGCAGC CTCATTCTCCTGCTCCTTCTCC |
| BiP | TGTTCAACCAATTATCAGCAAACTC TTCTGCTGTATCCTCTTCACCAGT |
TTCAGCCAATTATCAGCAAACTCT TTTTCTGATGTATCCTCTTCACCAGT |
TGGGTACATTTGATCTGACTGGA CTCAAAGGTGACTTCAATCTGGG |
| GRP94 | GAAACGGATGCCTGGTGG GCCCCTTCTTCCTGGGTC |
AAGAATGAAGGAAAAACAGGACAAAA CAAATGGAGAAGATTCCGCC |
|
| EDEM | CAAGTGTGGGTACGCCACG AAAGAAGCTCTCCATCCGGTC |
CTACCTGCGAAGAGGCCG GTTCATGAGCTGCCCACTGA |
As stated above, a 26-nucleotide intron of XBP-1 mRNA is spliced out under ER stress conditions leading to a frame shift. Taking advantage of the ER stress-dependent splicing of XBP-1, a fluorescent signal-based ER stress reporter has been developed [46]. A gene encoding venus, a variant of green fluorescent protein, is fused to human XBP1 downstream of the 26-nt intron. Under normal conditions, the mRNA of this fusion gene is not spliced, and therefore its translation is terminated at a stop codon between XBP1 and venus genes. However, under ER stress conditions, the 26-nt intron is spliced out, leading to a frameshift allowing the production of XBP1-venus fusion protein which can be detected by monitoring the fluorescence activity of venus. In transgenic mice expressing XBP-1-venus fusion protein, strong fluorescence in the kidney and pancreas was detected when tunicamycin was injected intraperitoneally. Thus, transgenic mice expressing XBP-1-venus could be used to monitor ER stress levels in vivo.
IRE1α knockout mouse embryonic fibroblasts are available in the research community [7, 10, 47]. These cells can be used as a negative control for assessing activation levels of IRE1α and its downstream components.
5.2. Methods for measuring PERK activation
Similar to IRE1α, PERK also undergoes transautophosphorylation upon ER stress. Phosophorylation levels of PERK can be detected by a phospho-specific PERK antibody (Cell Signaling Technologies, Danvers, MA). Upon activation, PERK phosphorylates eIF2α to reduce global mRNA translation. Measuring eIF2α phosphorylation levels by immunoblot using anti-phopho-eIF2α specific antibody (Cell Signaling, Danvers, MA) indirectly reflects PERK activation. However it must be noted that other eIF2α kinases exist and therefore proper controls should be included to confirm PERK dependent eIF2α phosphorylation.
Salubrinal is a selective inhibitor of cellular complexes that dephosphorylate eIF2α [48]. Thus, addition of salubrinal to cells enhances eIF2α phosphorylation. This compound can be used to study the eIF2α-dependent arm of PERK signaling. We treat cells with 75 µM salubrinal for 16 hours to observe enhanced eIF2α phosphorylation.
Other available tools include PERK knockout mice and PERK knock out mouse embryonic fibroblasts [21, 49, 50].
5.3. Methods for measuring ATF6α activation
As mentioned previously, in response to ER stress, ATF6α (90 kDa) transits to the Golgi apparatus and cleaved by SP1 and S2P producing a 50 kDa form which translocates to the nucleus to activate transcription of UPR genes [18]. By transfecting cells with a GFP-ATF6α fusion protein, ATF6 translocation events upon ER stress can be monitored by fluorescence microscropy. [51] Detection of the cleaved 50-kDa form of ATF6α by immunoblot using anti-ATF6α specific antibody (Imgenex) can be used as an indicator of ATF6α activation. The immunoblot protocol includes an unmasking step after blocking in order to reveal the antigen for antibody binding. Incubate the membrane in a sealed ziplock bag containing 50 ml of unmasking buffer (2% w/v SDS, 62.5mM TrisHCl or standard 1x PBS, 100 mM β-mercaptoethanol) in a 70°C water bath for 30 min. Discard the unmasking buffer and wash blot twice with PBS or Tris-HCl. Reblock the membrane and continue the protocol as usual. Another alternative to study ATF6α activation is measuring mRNA expression levels of genes that are regulated transcriptionally by ATF6α. Details of measuring transcription will be discussed later on.
6. Studying UPR downstream markers and responses
6.1. Immunostaining and immunofluorescence for downstream markers of the UPR
Immunostaining can also be used to measure UPR activation. [CO4]The advantage of immunostaining is that we can study tissues from patients or mouse models with ER stress-related diseases. Many of the antibodies discussed previously could be used for immunocytochemistry. In addition, CHOP, BiP and PDI antibodies can be used as indicators of cells undergoing ER stress conditions. CHOP is regulated under the PERK-eIF2α-ATF4 pathway and has been shown to have a role in ER stress mediated apoptosis. However, it must be cautioned that many commercially available antibodies for detection of CHOP expression fail specificity evaluation [52]. BiP is a central regulator of the UPR stress sensors as well as an ER chaperone to assist protein folding. BiP is highly expressed in the ER and can be used as an ER marker. PDI is involved in oxidative protein folding in the ER lumen and its expression is induced by ER stress.
To perform immunocytochemistry, we grow our cells typically onto four well Lab-Tek chambers. After ER stress induction, cells are fixed with 4% paraformaldehyde in PBS for 30 min at room temperature followed by permeabilization with 4% paraformaldehyde and 0.1 % Triton for 2 min at room temperature. Chambers are removed and slides are rinsed in PBST (PBS + 0.1% Tween). Non-specific binding sites are blocked by incubating samples with Image-iT Signal Enhancer (Invitrogen) for 30 min at room temperature. Samples are incubated with diluted [CO5]primary antibody in antibody diluent (Dako) overnight at 4°C in a humidified chamber. The next day slides are rinsed with PBST three times and then incubated with diluted secondary antibody in antibody diluent (1:200 – 1:1000) for 1 hour at room temperature. Slides are rinsed four times with PBST and one more time with PBS. Finally slides are mounted with ProLong Gold antifade mounting medium containing DAPI to stain DNA (Molecular Probes, Invitrogen). Slides could be viewed immediately by fluorescence microscopy or stored in the dark at 4°C for a month and at −80°C for several months.
6.2. Measuring transcriptional activation of the UPR
Upon ER stress conditions, activated master regulators of the UPR communicate to the nucleus to regulate the transcription of genes involved in protein folding and processing to increase the ER protein folding capacity, ERAD and autophagy components to reduce the ER workload, and cell survival and death factors to determine the fate of the cell depending on the ER stress condition.
IRE1α, PERK and ATF6α are all involved in regulating transcription during ER stress. IRE1α directly regulates the splicing of XBP1 mRNA to produce a transcriptionally active basic leuzine zipper transcription factor. XBP1 regulates chaperones, folding catalysts and ERAD components such as BiP, EDEM, and HRD1. Phosphorylated eIF2α by PERK generally reduces mRNA translation; however, preferentially favors translation of some mRNAs such as the transcription factor ATF4. ATF4 regulates genes involved in antioxidative stress, amino acid biosynthesis, protein folding and degradation, and apoptosis such as C/EBP homologous protein (CHOP). CHOP of GADD153 is a bZIP transcription factor regulating apoptosis related genes such as death receptor 5 (DR5), tribble 3 (TRB3), and members of the BCL2 family of proteins. One of the most reliable methods to measure transcriptional regulation of the UPR in ER stressed cells is by quantitative real-time PCR. Table 2 lists commonly measured UPR genes and their primers.
Many of the genes regulated by the UPR contain unique cis-acting response elements within their promoters. These include ERSE (ER stress response element, 5'-CCAAT-N9-CCACG-3'), ERSE-II (ER stress response element II, 5’-ATTGG-N1-CCACG-3’) and the UPRE (Unfolded Protein Response element, 5’-TGACGTGG/A-3’).
Instead of measuring mRNA expression levels of UPR genes, there are several reporter systems that reflect endogenous UPR activation levels.[CO6] A luciferase plasmid driven by the human Grp78 promoter is commonly used. The promoter contains three copies of ERSE upstream of the TATA element. Another luciferase reporter often studied contain one or five ATF6α binding sites[53]. This reporter can be activated by ER stress inducers as well as ATF6α and XBP-1 overexpression. Cells are transfected with luciferase reporters, overexpression vectors, and beta galactosidase internal control using optimized transfection methods. We commonly transfect COS7 or 293T cells by Lipofectamine 2000 (Invitrogen) for luciferase assays. After 24–48 hours post-transfection, cells are treated with ER stress inducers and/or harvested using the Luciferase Assay System kit (Promega). We have determined that low dose ER stress (e.g. 50 nM Tg and 0.5 ug/ml tunicamycin, 18hours) can activate GRP78 and ATF6α luciferase reporters. Firefly luciferase and beta galactisade activitities (β-gal reporter gene assay, chemiluminescent, Roche) are measured by a standard plate reading luminometer.
6.3. Measuring translational attenuation of the UPR
Translational attenuation is an early UPR response in order to reduce the ER protein workload. In response to ER stress, cells attenuate translation through PERK-mediated eIF2α phosphorylation. As mentioned earlier, eIF2α phosphorylation can be detected by western blot. Translational attenuation can be measured by metabolic pulse labeling of newly synthesized proteins and polyribsome profiling using standard protocols [13]. As the UPR restores ER homeostasis, GADD34 interacts with protein phosphatase 1c to dephosphorylate eIF2α restoring protein synthesis. [54] Gadd34 expression is induced by ER stress and regulated under the PERK-ATF4 pathway.
6.4. Measuring ERAD and protein stability
The UPR removes harmful proteins by regulating expression of ERAD genes. During ERAD, misfolded and unfolded proteins are recognized by ER chaperones, retrotranslocated out of the ER into the cytosol, and finally ubiquitinated and degraded by the proteasome. ATF6, XBP-1, and ATF4 are all involved in regulating the transcription of ERAD components such as EDEM and HRD1. To study the ERAD pathway, three proteins susceptible to misfolding in the ER; TCRα, mutant alpha-1-antitrypsin NHK3 and the DeltaF508-variant cystic fibrosis transmembrane conductance regulator protein, are often used [55–58]. These substrates can be ectopically expressed in cells and their stability can be monitored by cycloheximide chase or metabolic pulse-chase labeling assays. [CO8]Degradation rates of these proteins reflect the activation levels of ERAD in cells. In addition ubiquitination of an interested ERAD substrate can be studied by treating cells with the proteasome inhibitor, MG132, immoprecipitating the protein of interest followed by immunoblot with anti-ubiquitin antibody (Cell signaling).
6.5. Measuring mRNA degradation
It has been shown that ER stress accelerates degradation of mRNAs in cells [59]. This is largely dependent on the RNase activity of IRE1α [59, 60]. Under unresolvable ER stress conditions, the RNase domain of IRE1α plays a role in degrading mRNAs encoding secretory proteins in addition to splicing XBP-1 mRNA. In β cell lines, insulin mRNA has been shown to be a substrate of IRE1 and is quickly degraded under ER stress conditions [61–63]. This phenomenon can be used in detecting and quantifying unresolvable ER stress in pancreatic β cells. Cellular mRNA transcription is attenuated by treating β cells with 100µg/mL actinomycin D for 1 hr. Total RNA is isolated at different time points, reverse transcribed to cDNA, and insulin gene transcripts are measured by real-time PCR as described before. Time point zero for each condition is standardized to 1 and the subsequent rate of degradation of mRNA is measured. Degradation rate of insulin mRNA could be used as a biomarker for β cells experiencing unresolvable ER stress.
6.6. Measuring ER stress-mediated apoptosis
When the UPR fails to restore ER homeostasis and attenuate ER stress, the UPR activation induces apoptosis. ER stress-mediated apoptosis is involved in many human chronic diseases. Thus, measuring ER stress-mediated apoptosis will aid us in understanding the pathogenesis of ER stress-related disorders.
There are several components of the UPR that could contribute to ER stress-mediated apoptosis including the IRE1α-ASK1-JNK signaling pathways, CHOP regulation of BCL2 protein family members and apoptotic genes, ER localized Bax and Bak, and glycogen synthase kinase 3β (GSK3β). Activated IRE1α binds to the adaptor protein TRAF2 and subsequently activate ASK1, which activates the JNK pathway. The JNK pathway has been shown to play an important role in ER stress-mediated cell death by regulating the BCL2 family of proteins [10, 11, 64]. IRE1α, ASK1, and JNK are serine/threonine protein kinases and therefore their activation levels can be measured by phospho-specific antibodies.[CO10] CHOP is a pro-apoptotic transcription factor of the UPR [65]. Because its baseline expression is low, its upregulation and activation can be measured by immunoblot or real-time PCR as mentioned previously. Cells undergoing ER stress mediated cell death can also be determined by immunostaining for CHOP. Proapoptotic BCL-2 family members BAX and BAK are associated with the ER membrane. Upon ER stress, BAK and BAK undergo conformational changes forming pores in the membrane causing Ca2+ to leak into the cytosol, which in turn stimulate the activation of apoptotic pathways. BAX and BAK double knockout cells are resistant to ER stress-mediated cell death [66, 67]. BAX and BAK expression levels can be detected by immunoblot. [CO11]Finally GSK3β also plays a role in ER stress-mediated apoptosis. GSK3β is a substrate of the survival kinase, Akt [68], and it has been demonstrated that attenuation of Akt phosphorylation during ER stress mediates dephosphorylation of GSK3β, leading to ER stress-mediated apoptosis [69]. GSK3β phosophorylation levels can be measure by anti-phospho-specific GSK3β antibody.
ER stress-mediated apoptosis can be measured by standard methods. Measuring caspase-3 cleavage (Cell Signaling) by immunoblot, staining the cells with PE Annexin-V (BD Biosciences) followed by FACS analysis, TUNEL staining using the DeadEnd™ Colorimetric TUNEL System (Promega), and cell viability assays using CellTiter-Glo (Promega) are commonly used in our lab.
Acknowledgments
This work was supported in part by grants from NIH-NIDDK (R01DK067493), the Diabetes and Endocrinology Research Center at the University of Massachusetts Medical School (5 P30 DK32520), and the Juvenile Diabetes Research Foundation International to F. Urano.
References
- 1.Kim I, Xu W, Reed JC. Nat Rev Drug Discov. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 2.Ron D, Walter P. Nature reviews. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 3.Oslowski CM, Urano F. Curr Opin Endocrinol Diabetes Obes. 2010 doi: 10.1097/MED.0b013e3283372843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urano F, Bertolotti A, Ron D. Journal of cell science. 2000;113:3697–3702. doi: 10.1242/jcs.113.21.3697. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 6.Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 7.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. Dev Cell. 2003;4:265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 9.Lee AH, Iwakoshi NN, Glimcher LH. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Science (New York, N.Y. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 11.Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K, Hori S, Kakizuka A, Ichijo H. Genes Dev. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishitoh H, Saitoh M, Mochida Y, Takeda K, Nakano H, Rothe M, Miyazono K, Ichijo H. Molecular Cell. 1998;2:389–395. doi: 10.1016/s1097-2765(00)80283-x. [DOI] [PubMed] [Google Scholar]
- 13.Harding HP, Zhang Y, Ron D. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 14.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 15.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. The Journal of biological chemistry. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 17.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. Mol Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertolotti A, Wang X, Novoa I, Jungreis R, Schlessinger K, Cho JH, West AB, Ron D. Journal of Clinical Investigation. 2001;107:585–593. doi: 10.1172/JCI11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Molecular Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 22.Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 24.Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, Grusby MJ, Glimcher LH. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- 25.Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 26.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 27.Novoa I, Zhang Y, Zeng H, Jungreis R, Harding HP, Ron D. Embo J. 2003;22:1180–1187. doi: 10.1093/emboj/cdg112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyazaki J, Araki K, Yamato E, Ikegami H, Asano T, Shibasaki Y, Oka Y, Yamamura K. Endocrinology. 1990;127:126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 29.Asfari M, Janjic D, Meda P, Li G, Halban PA, Wollheim CB. Endocrinology. 1992;130:167–178. doi: 10.1210/endo.130.1.1370150. [DOI] [PubMed] [Google Scholar]
- 30.Hohmeier HE, Mulder H, Chen G, Henkel-Rieger R, Prentki M, Newgard CB. Diabetes. 2000;49:424–430. doi: 10.2337/diabetes.49.3.424. [DOI] [PubMed] [Google Scholar]
- 31.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cardozo AK, Ortis F, Storling J, Feng YM, Rasschaert J, Tonnesen M, Van Eylen F, Mandrup-Poulsen T, Herchuelz A, Eizirik DL. Diabetes. 2005;54:452–461. doi: 10.2337/diabetes.54.2.452. [DOI] [PubMed] [Google Scholar]
- 33.Kharroubi I, Ladriere L, Cardozo AK, Dogusan Z, Cnop M, Eizirik DL. Endocrinology. 2004;145:5087–5096. doi: 10.1210/en.2004-0478. [DOI] [PubMed] [Google Scholar]
- 34.Oyadomari S, Koizumi A, Takeda K, Gotoh T, Akira S, Araki E, Mori M. Journal of Clinical Investigation. 2002;109:525–532. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng B, Yao PM, Li Y, Devlin CM, Zhang D, Harding HP, Sweeney M, Rong JX, Kuriakose G, Fisher EA, Marks AR, Ron D, Tabas I. Nature cell biology. 2003;5:781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- 36.Outinen PA, Sood SK, Pfeifer SI, Pamidi S, Podor TJ, Li J, Weitz JI, Austin RC. Blood. 1999;94:959–967. [PubMed] [Google Scholar]
- 37.Riggs AC, Bernal-Mizrachi E, Ohsugi M, Wasson J, Fatrai S, Welling C, Murray J, Schmidt RE, Herrera PL, Permutt MA. Diabetologia. 2005;48:2313–2321. doi: 10.1007/s00125-005-1947-4. [DOI] [PubMed] [Google Scholar]
- 38.Akiyama M, Hatanaka M, Ohta Y, Ueda K, Yanai A, Uehara Y, Tanabe K, Tsuru M, Miyazaki M, Saeki S, Saito T, Shinoda K, Oka Y, Tanizawa Y. Diabetologia. 2009;52:653–663. doi: 10.1007/s00125-009-1270-6. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Takeuchi T, Tanaka S, Kubo SK, Kayo T, Lu D, Takata K, Koizumi A, Izumi T. Journal of Clinical Investigation. 1999;103:27–37. doi: 10.1172/JCI4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tu BP, Weissman JS. J Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 42.Merksamer PI, Trusina A, Papa FR. Cell. 2008;135:933–947. doi: 10.1016/j.cell.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen JR, Nguyen LX, Sargent KEG, Lipson KL, Hackett A, Urano F. Biochem Biophys Res Commun. 2004;324:166–170. doi: 10.1016/j.bbrc.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 44.Fonseca SG, Ishigaki S, Oslowski CM, Lu S, Lipson KL, Ghosh R, Hayashi E, Ishihara H, Oka Y, Permutt MA, Urano F. The Journal of clinical investigation. 2010 doi: 10.1172/JCI39678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishigaki S, Fonseca SG, Oslowski CM, Jurczyk A, Shearstone JR, Zhu LJ, Permutt MA, Greiner DL, Bortell R, Urano F. Cell death and differentiation. 2010;17:774–786. doi: 10.1038/cdd.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwawaki T, Akai R, Kohno K, Miura M. Nat Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- 47.Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The Journal of clinical investigation. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, Yuan J. Science (New York, N.Y. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 49.Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR. Molecular & Cellular Biology. 2002;22:3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang W, Feng D, Li Y, Iida K, McGrath B, Cavener DR. Cell Metab. 2006;4:491–497. doi: 10.1016/j.cmet.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 51.Nadanaka S, Yoshida H, Kano F, Murata M, Mori K. Mol Biol Cell. 2004;15:2537–2548. doi: 10.1091/mbc.E03-09-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haataja L, Gurlo T, Huang CJ, Butler PC. Cell Biochem Biophys. 2008;51:105–107. doi: 10.1007/s12013-008-9019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Journal of Biological Chemistry. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- 54.Novoa I, Zeng H, Harding HP, Ron D. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu H, Kaung G, Kobayashi S, Kopito RR. The Journal of biological chemistry. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- 56.Yu H, Kopito RR. The Journal of biological chemistry. 1999;274:36852–36858. doi: 10.1074/jbc.274.52.36852. [DOI] [PubMed] [Google Scholar]
- 57.Hosokawa N, Tremblay LO, You Z, Herscovics A, Wada I, Nagata K. The Journal of biological chemistry. 2003;278:26287–26294. doi: 10.1074/jbc.M303395200. [DOI] [PubMed] [Google Scholar]
- 58.Hosokawa N, Wada I, Hasegawa K, Yorihuzi T, Tremblay LO, Herscovics A, Nagata K. EMBO Rep. 2001;2:415–422. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hollien J, Weissman JS. Science (New York, N.Y. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 60.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lipson KL, Ghosh R, Urano F. PLoS ONE. 2008;3:e1648. doi: 10.1371/journal.pone.0001648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pirot P, Naamane N, Libert F, Magnusson NE, Orntoft TF, Cardozo AK, Eizirik DL. Diabetologia. 2007;50:1006–1014. doi: 10.1007/s00125-007-0609-0. [DOI] [PubMed] [Google Scholar]
- 64.Nishitoh H, Kadowaki H, Nagai A, Maruyama T, Yokota T, Fukutomi H, Noguchi T, Matsuzawa A, Takeda K, Ichijo H. Genes Dev. 2008;22:1451–1464. doi: 10.1101/gad.1640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zong WX, Lindsten T, Ross AJ, MacGregor GR, Thompson CB. Genes Dev. 2001;15:1481–1486. doi: 10.1101/gad.897601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zong WX, Li C, Hatzivassiliou G, Lindsten T, Yu QC, Yuan J, Thompson CB. J Cell Biol. 2003;162:59–69. doi: 10.1083/jcb.200302084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 69.Srinivasan S, Ohsugi M, Liu Z, Fatrai S, Bernal-Mizrachi E, Permutt MA. Diabetes. 2005;54:968–975. doi: 10.2337/diabetes.54.4.968. [DOI] [PubMed] [Google Scholar]