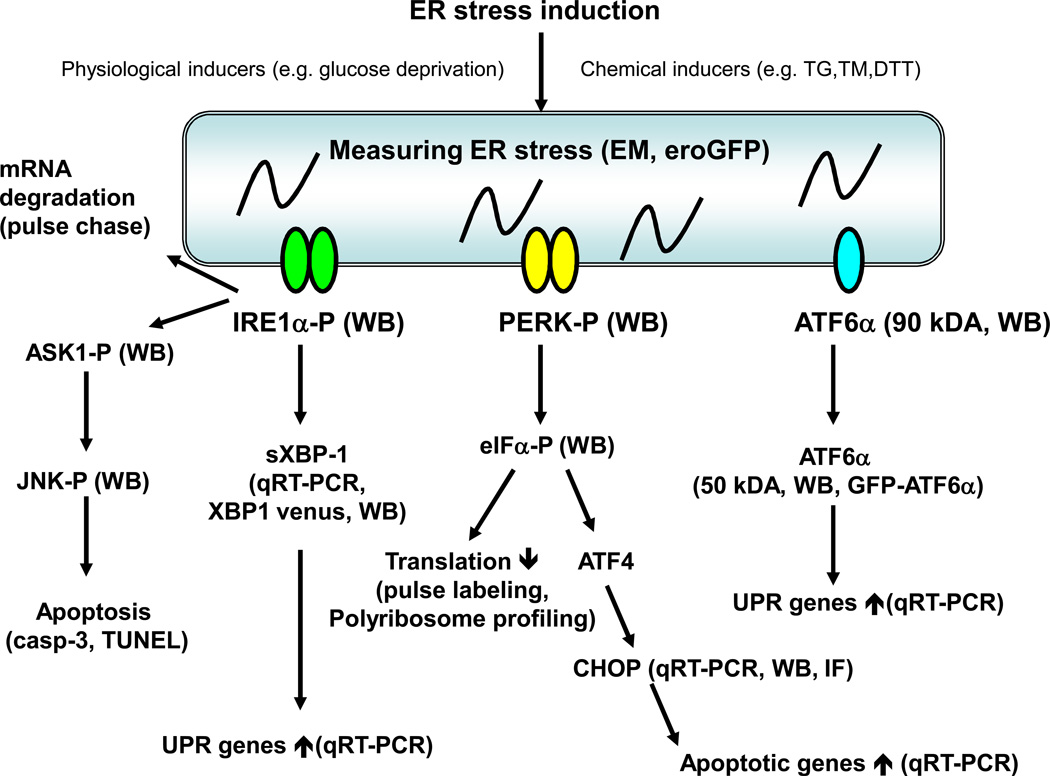
Figure 2. Studying ER stress and the Unfolded Protein Response in Mammalian cells.
ER stress can be induced by several chemical and physiological inducers. Actual ER stress has been difficult to measure directly. Currently observing ER distension by electron microscopy (EM) and measuring oxidative protein folding (eroGFP) are available. The activation of the UPR master regulators has also been challenging but could be attempted by detecting IRE1α and PERK phosphorylation, and ATF6α cleavage by specific antibodies using western blot (WB). ATF6α translocation can be monitored by fluorescence microscopy of GFP-ATF6α. The downstream outputs of the UPR master regulars are readily measurable. IRE1α splices XBP-1 mRNA which can be detected by quantitative real time PCR (qRT-PCR), XBP-1 venus, and western blot (WB). IRE1α also mediates degradation of ER localized mRNAs, which can be measured by pulse chase assays. PERK phosphorylates eIF2α, which can be detected by specific antibodies. Phosphorylated eIF2α triggers global mRNA translation attenuation, which can be measured by standard methods such as pulse labeling and polyribsome profiling. Activated ATF6α is a transcription factor and regulation of its downstream target genes can be measured by qRT-PCR. The UPR in general regulates several transcription factors and in turn their transcriptional targets can be measured by qRT-PCR and luciferase assays. The UPR also induces expression of ERAD components as well as survival and death components. ERAD and apoptosis can be measured by standard methods.