Fig. 2.
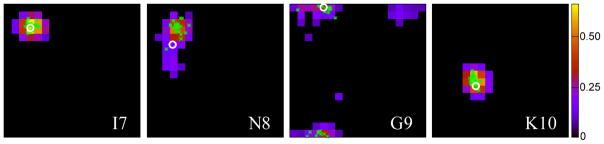
(φ,ψ)-ANN predicted (φ,ψ) likelihood distributions presented as Ramachandran maps for residues 7 to 10 of protein GB3. Only 20°×20° voxels with predicted likelihoods that fall at least one standard deviation above the average population (1/324) are color coded. The φ/ψ angles observed in the reference structure (first conformer of PDB entry 2OED) is marked with a white circle; the φ/ψ angles of the center residue of the 25 best matched database fragments are displayed as green dots. The horizontal axis of each plot corresponds to φ (ranging from −180° to 180°) with the vertical axis being ψ (ranging from −180° to 180°, bottom to top). Residue G9 shows two clusters; one centered near (φ,ψ) = (−100°, 180°) and one at (120°, 160°).
