Figure 2 .
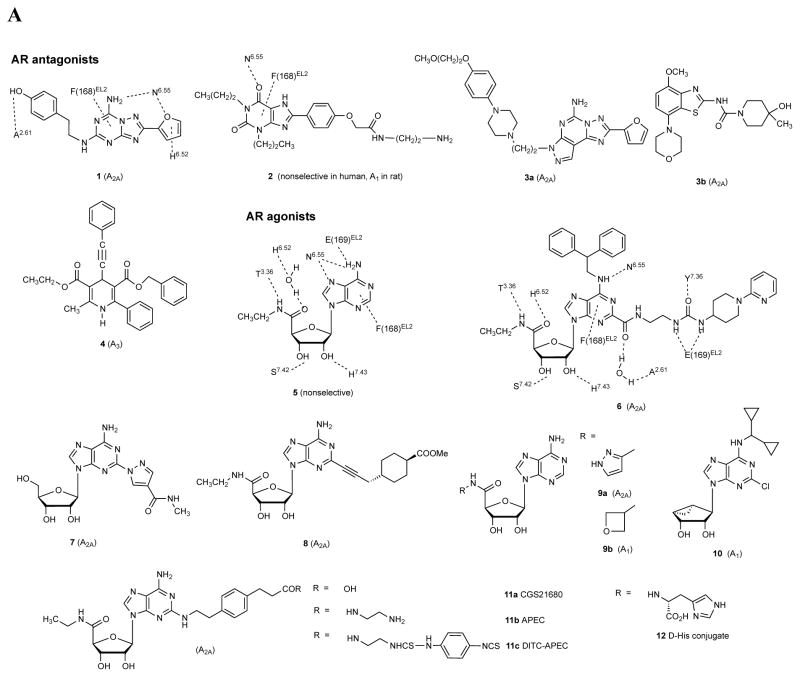
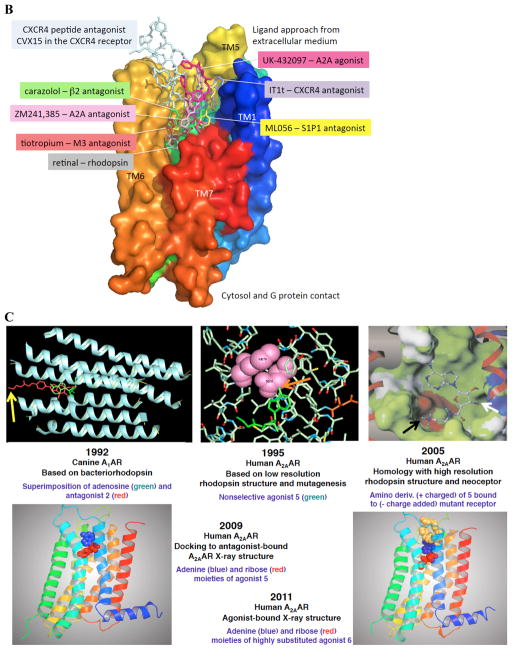
A. Structures of selected AR ligands discussed in the text, including those that have been co-crystallized with the A 2A AR ( 1 , 2 , 5 , and 6 ). Dashed lines indicated the major H-bonding and π-bonding contacts between the receptor and ligand present in X-ray structures. Other ligands shown include pharmacological probes ( 4 and 9–12 ) and: advanced clinical candidates 3a and 3b for Parkinson’s disease; diagnostic agents for myocardial perfusion imaging 7 (approved and in trials for sickle cell anemia and ischemic conditions) and 8 (clinical candidate). B. The helical bundle of Family A GPCRs defines a cavity for the recognition of diverse ligands. The phospholipid bilayer is not shown. Figure courtesy of Stefano Costanzi (American Univ., Washington, DC). C. Historical progression of knowledge of the AR binding site(s). 19 , 20 , 37 , 39 , 41 , 45 Arrows in upper panels indicate the following interactions: Yellow, position of terminal amino group intended for covalent linking 37 ; orange, H-bonding interaction predicted between the exocyclic amine of adenine and a conserved Asn6.55 39 ; white, proximity of 3′ and 2′-hydroxyl groups with conserved His7.43 predicted using a neoceptor approach 41 . Lower panels: left, predicted docking of agonist 5 using the antagonist-bound X-ray structure 19 , 45 ; right, actual position of agonist 6 in X-ray structure. 20 , 24