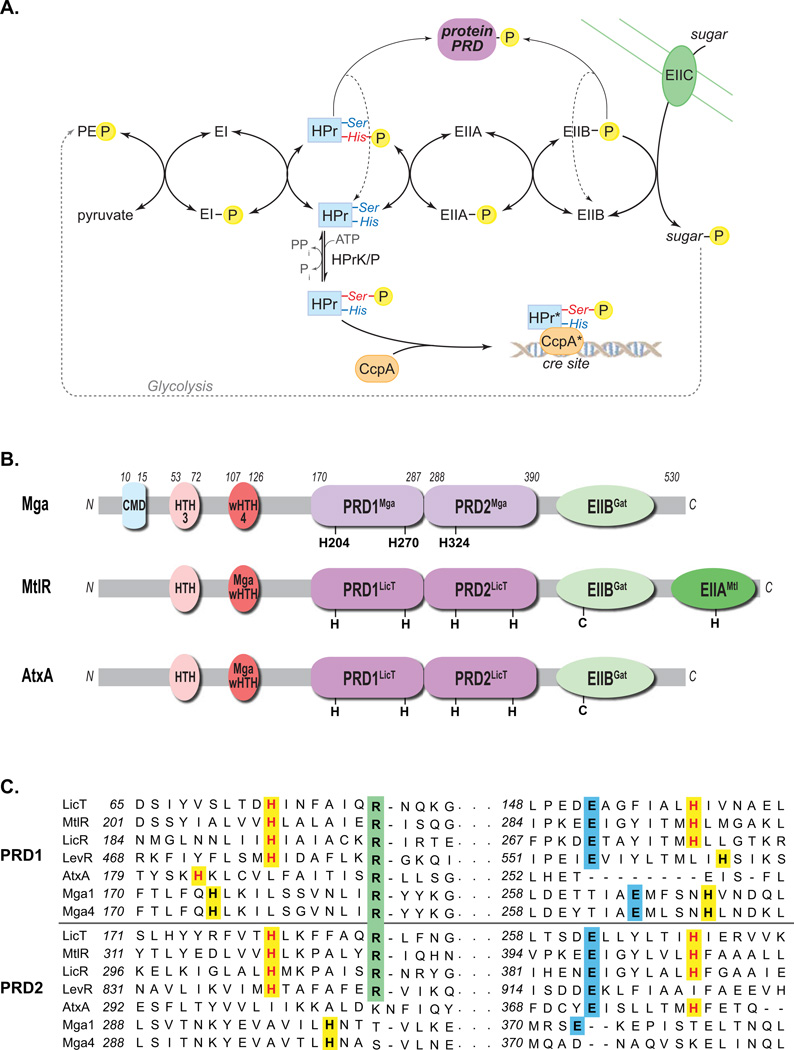
Figure 1. PTS pathway and alignment of Mga with PRD-containing regulators.
(A) The phosphotransferase system (PTS) in Gram-positive bacteria couples the phosphorylation and import of sugars. The general cytoplasmic enzymes (EI and HPr) and sugar-specific EII components form a phosphorelay to transfer phosphate from phosphoenol pyruvate (PEP) produced by glycolysis to the incoming sugar. Carbon catabolite repression (CCR) results from HPrK/P phosphorylation of serine 46 of HPr, which then complexes with CcpA to repress target promoters via cre sites. HPr-His∼P and EIIB∼P can also phosphorylate PTS regulatory domains (PRDs) of sugar regulators, thereby modulating their activity. (B) Domain alignment of GAS Mga with B. subtilis MtlR and B. anthracis AtxA. EII (green), PRD (purple), DNA-binding (red) and conserved (blue) domains are indicated with conserved histidines (H) and cysteines (C). PRDMga and PRDLicT refer to Pfam 08270 and Pfam 00874, respectively. (C) Amino acid sequence alignment of the PRDs from B. subtilis LicT, LicR, LevR, G. stearothermophilus MtlR, B. anthracis AtxA, and GAS Mga (M1 and M4 serotypes), highlighting the regions containing the conserved histidine (highlighted yellow), arginine (highlighted green), and aspartate (highlighted blue) residues. Histidine residues previously shown to be phosphorylated are colored red. (Supplemental Figure S1 contains the full alignment.)