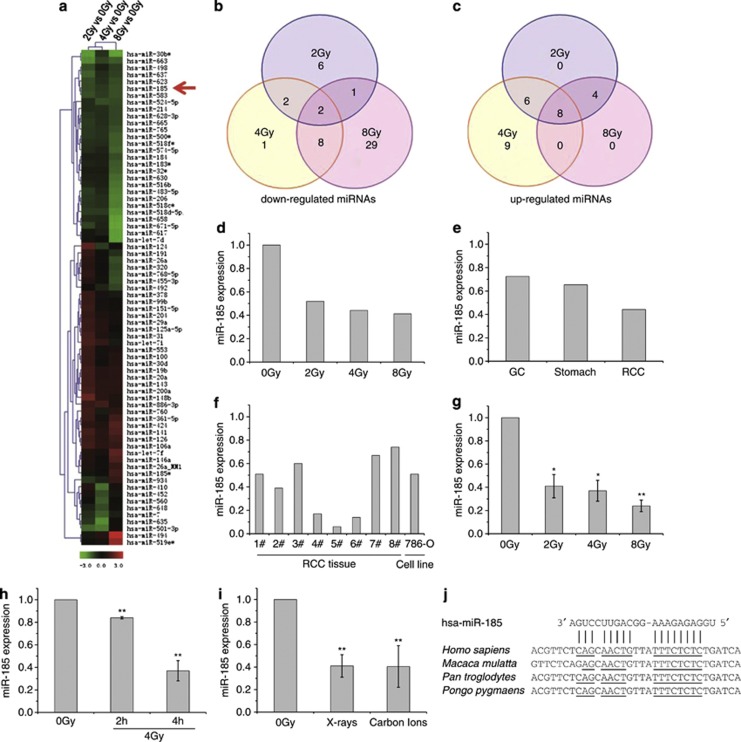
Figure 1.
miR-185 expression is downregulated in response to ionizing radiation. (a) Hierarchical clustering of miRNA expression obtained with Exiqon miRNA microarrays (version 9.2) after exposure of clinical RCC tissues to 2, 4 and 8 Gy of X-rays, respectively. Colored bars represent the differential levels of miRNAs expressed in irradiated samples versus sham-irradiated samples (0 Gy). The red arrow indicates hsa-miR-185. (b) Venn diagram of the miRNA expression profile depicting consistency of downregulated miRNAs between samples. (c) Venn diagram of the miRNA expression profile depicting consistency of upregulated miRNAs between samples. (d) Relative changes of the miR-185 expression level in clinical RCC tissues 4 h after X-ray irradiation. (e) Relative changes of the miR-185 expression level in various clinical tissues 4 h after 4 Gy X-ray irradiation, which was also obtained with a miRNA microarray assay. GC, gastric cancer tissues; Stomach, normal stomach tissues. (f) Verification of miR-185 expression altered by 4 Gy of X-rays in clinical RCC tissues obtained from patients (n=8) and in the cultured RCC cell line 786-O, analyzed by qRT-PCR. The histograms show the relative changes of the miR-185 expression level compared with sham-irradiated samples (0 Gy). (g) Dose-dependent inhibition of miR-185 expression in 786-O cells 4 h after irradiation, obtained by qRT-PCR. (h) Time-dependent inhibition of miR-185 expression in 786-O cells exposed to 4 Gy of X-rays, obtained by qRT-PCR. (i) Radiation-type-dependent inhibition of miR-185 in 786-O cells exposed to 2 Gy X-rays or carbon ions, obtained by qRT-PCR. (j) Predicted binding sites of miR-185 on the 3′-UTR of ATR mRNA in several species. Data with error bars represent the means of at least three independent experiments while others represent the means of two independent experiments. *P<0.05 compared with the sham-irradiated samples (0 Gy); **P<0.01 compared with the sham-irradiated samples (0 Gy)