All the crystallization communications published in Acta Cryst. F in 2012 were analysed. Details of the analysis are presented, along with some suggestions for making this type of publication more useful.
Keywords: crystallization, crystallization communications
Abstract
Crystallization of macromolecules is famously difficult. By knowing what has worked for others, researchers can ease the process, both in the case where the protein has already been crystallized and in the situation where more general guidelines are needed. The 264 crystallization communications published in Acta Crystallographica Section F in 2012 have been reviewed, and from this analysis some information about trends in crystallization has been gleaned. More importantly, it was found that there are several ways in which the utility of these communications could be increased: to make each individual paper a more complete crystallization record; and to provide a means for taking a snapshot of what the current ‘best practices’ are in the field.
1. Introduction
The realities of science funding today mean that the goal of a researcher cannot simply be to investigate interesting scientific questions. Without performing well on the metrics that are used to judge eligibility for support (e.g. government research grants), researchers simply will not (or will cease to) attract funding. A primary metric is the h-index or some variant (Bornmann et al., 2008 ▶) i.e. the number of papers published by an author that have received more than h citations. Thus, a driving force when preparing a manuscript is to produce as high an impact paper as possible. Initially, this comes from publishing in high-impact journals (which are more likely to be read) and then over time the citation count of the paper becomes a more realistic validation of the impact of the work. In an ideal world, every publication should provide enough detail of the experiments presented therein so that anyone could reproduce the experiments and convince themselves of the results and conclusions presented.
Of course, it is unlikely that anyone has the resources to reproduce all the interesting results from the literature, so this absolute requirement can slip without causing a great outcry. Indeed, a recent commentary in Nature shows just how unlikely it is that even those skilled in the art can now reproduce an experiment (Begley & Ellis, 2012 ▶). Compounding this is the ever-increasing trend of journals to limit the amount of experimental detail that can be included in a research paper. Often, supporting information (generally the Materials and methods section) is relegated to a second tier, associated document: the supplementary information. Supplementary information has many issues, not the least being that any citations in supplements are not counted (Seeber, 2008 ▶; Weiss et al., 2010 ▶).
We are long past the time when every crystal structure is a novelty; the typical structural biologist uses structure as a tool to understand the biology of a system, and thus the resulting papers emphasize that. Of course there are exceptions: the recent work using the free electron lasers in very high profile journals attests to this (see Boutet et al., 2012 ▶; Chapman et al., 2011 ▶). It is also a truism that the higher impact the journal, the less space that is devoted to the experimental details. Given that the production of the sample and its crystallization are the known bottlenecks in the production of structures, what is the best way to enable the widespread dissemination about the current best practices in these areas?
One way is to have specialist publications which describe in detail the process of producing the sample, not just the final crystallization experiment that produced the crystal used in the diffraction analysis which is what is found in most Materials and methods sections. These papers would include information about producing and storing the sample, the initial screening for likely conditions, and then the process of optimization, leading ultimately to an unambiguous description of the experiment that ultimately yielded the diffraction quality crystals. Indeed, there is a class of publications that are devoted to this information: these papers are often entitled some variation of The crystallization and preliminary X-ray analysis of … In the past, this type of information was readily publishable in quite mainstream journals, and would often work as a claim stake for a particular area of biology. Today, these types of papers do not seem to be highly valued: more often than not they get at most one citation, from the paper describing the associated X-ray structure.
Despite doing poorly on arbitrary metrics, crystallization papers can be very worthwhile. First and foremost, the perfect crystallization paper should allow a skilled researcher to replicate the crystals, and provide information about pitfalls that might be encountered replicating the work, and details about the crystals that might allow an informed decision about using them for further work, for example, compound binding studies. Secondly, these papers should (collectively) give a snapshot of crystallization technology and current best practices. Thirdly, these papers provide a forum for tips or techniques that could be more broadly applied. Additionally these papers are useful as teaching tools: they are very structured papers which provide students and early career scientists with a good introduction to the craft of scientific communication, from the perspective of both writing and reviewing.
Much of the information in the crystallization communications is useful en masse, for example, finding out which of the multitude of commercially available screens is currently proving to be most successful. To find this information requires (at least) two prerequisites: that the information about which screen(s) were tested is recorded, and that the relevant literature can be searched effectively to extract the information. We were interested in finding out what types of systems were being described in the crystallization papers published in Acta F in 2012, as well as discovering if there are changes that could be made to the process of writing, editing and publishing these communications with the goal of making these papers more robust, more relevant and thus more useful to the structural biology community (and incidentally, more cited) than they currently are.
2. Methods
A survey form was created (http://www.SurveyMonkey.com) which consisted of 29 questions, mostly multiple-choice questions, but some that required a text response (for example, ‘What was the final crystallization condition?’). The form was filled in at least once for each of the 264 crystallization communications published in 2012, more often if more than one ‘final’ crystallization condition was reported in the paper. Twelve of the authors of this communication were responsible for completing a survey for the crystallization communications for a calendar month, along with a single paper from the previous month and the following month. The duplicated surveys were used to provide some indication of how reliable the responses were from each author. There were three parts to the online survey.
(1) Initial questions that were used to identify the surveyor, the paper, and if the paper was one of the ‘in month’ or ‘out of month’ papers.
(2) The major part of the survey was dedicated to teasing out the details of each paper, and consisted of questions such as ‘How was the molecule purified? (Click all that apply)’ (followed by a list of likely purification techniques).
(3) The last questions were devoted to answering the question ‘Did the paper fulfill its primary goal?’ i.e. that of detailing unambiguously how to produce the crystals described therein. The final question allowed us to collect any noteworthy techniques that might be useful additions to the experimental arsenal of the crystal grower.
As the 12 people responsible for creating the surveys ranged from crystallographers with many years of experience in protein crystallization to early career PhD students, the questions were created with the aim of ensuring the extraction of information from the papers was as straightforward as possible; trying not to rely on the experience of the surveyor in order to glean information from the papers. To gauge the reliability of the surveying, the 24 ‘out of month’ papers were compared with the same papers surveyed as part of the ‘in month’ collections. As each surveyor had a paper from the previous month and the following month, each surveyor was compared to two others. To aid this analysis, a further online survey was created, where the questions related to the agreement of the answers given by the two surveyors to the questions of the first survey. Only one surveyor performed this overview analysis.
The consistency analysis suggests that there is some variation in the information that can be routinely obtained from crystallization communications, but some of the variation was undoubtedly owing to some ambiguity in the questions of the survey and the experience level of the surveyor. Overall, there was a high level of agreement found in the consistency analysis, generally over 80%.
The final set of crystallization conditions extracted from the papers was parsed firstly by a Python script then by hand for name consistency, and was subjected to an overall analysis akin to the ‘single screen statistics’ analysis available on the c6 web tool (Newman et al., 2010 ▶).
3. Results
Although there are other journals that accept crystallization papers, searching for ‘crystallization’ and ‘preliminary X-ray’ in Google Scholar (http://scholar.google.com) showed that Acta Crystallographica Section F appeared to be overwhelmingly the journal of choice for these types of dedicated crystallization papers in 2012, so the papers reviewed are potentially a large fraction of the specialist crystallization papers published in 2012. Given that there were 8321 X-ray structures deposited in the Protein Data Bank (PDB) in the same timeframe (http://www.rcsb.org/pdb/statistics/contentGrowthChart.do?content=explMethod-X-ray&seqid=100) it is clear that most structures do not have a dedicated crystallization paper associated with them. Of course, crystallization communications are by no means the only source of crystallization information (most structures are published with crystallization included in the materials and methods) Table 1 ▶ gives a (partial) list of journals, with the approximate number of structure papers found in each journal for 2012.
Table 1. This shows a selection of some of the more popular/high impact journals in which crystal structures are reported.
The number of structure papers, and by inference, the number of papers which would have some crystallization information associated with them, was estimated by using the search tools on the webpage for each journal, looking for all of the key words ‘crystal’, ‘structure’, ‘crystallization’ and ‘protein’ simultaneously, and limiting the search to articles published in the year 2012. The Cell family of journals includes Cancer Cell, Cell Host and Microbe, Cell Metabolism, Cell Stem Cell, Current Biology, Developmental Cell, Immunity, Molecular Cell and Neuron.
| Journal | No. of structure papers in 2012 |
|---|---|
| Journal of Biological Chemistry | 424 |
| Journal of Molecular Biology | 265 |
| Structure | 216 |
| Journal of Structural Biology | 153 |
| Acta Crystallographica Section D | 145 |
| Nature | 93 |
| Nature Structural Biology | 76 |
| Biochemistry | 74 |
| Proteins: Structure, Function and Bioinformatics | 52 |
| Cell family of journals | 50 |
| Science | 38 |
| Molecular Microbiology | 31 |
| Cell | 25 |
| EMBO Journal | 22 |
| PNAS | 19 |
3.1. Some trends in 2012
In 2012 the majority (75%) of Acta F crystallization communications reported the crystallization of a single protein chain, where most (>90%) of the protein samples were produced heterologously, most commonly (90%) in bacterial expression systems, using His tags (65%) located on the N-terminus (63%) of the protein. By far the most utilized purification techniques were affinity chromatography (75%) followed by size-exclusion chromatography (70%), with the next most frequently used purification technique being ion-exchange chromatography (40%). Most formulations contained 50–200 mM salt (53%), and 10–50 mM buffer (68%) at a pH between 7 and 8 (80%). Over 40% of the reported protein concentrations were between 10 and 20 mg ml−1, with 22% between 5 and 10 mg ml−1. The results for the characterization show that little characterization is reported; almost 60% of the papers reported using SDS–PAGE analysis, while the next most commonly used characterization technique reported, mass spectrometry, was seen in less than 15% of the papers. Despite a lot of emphasis on the predictive power of Dynamic Light Scattering (DLS) in the literature (see for example, Ferré-D’Amaré & Burley, 1997 ▶; Wilson, 2003 ▶) only 6% of papers reported using it as a characterization tool. Of the commercial screens explicitly named, Crystal Screen (117 papers) and Crystal Screen 2 (102 papers) from Hampton Research were most widely used. These were followed by Hampton’s Index Screen (88 papers), EmeraldBio’s Wizard Screen 1 and Wizard Screen 2 (33 papers each), Molecular Dimension’s Morpheus Screen (15 papers), and then PACT (28 papers) and JCSG+ (41 papers); these latter two screens were sourced from various vendors.
3.2. Not reported, or not reported well
Other questions were very difficult to answer from the papers, for example, in 70% of the papers, there was no indication if the protein was (or could be) frozen for long-term storage. A similar number of papers were reticent about the number of screening drops that were set up. The screening was poorly described both in methodology and outcome. 40% of the papers did not describe how many hits were obtained from the screening. In 22% of the papers only one hit was obtained, but from the papers it is unclear what constitutes a hit for the different research groups. Optimization of the screening hits was required 75% of the time, yet the description of the optimization tended to be quite unreliable, with over 95% of the papers not reporting how many optimization experiments were tried. Of the 240 surveys that reported that optimization was used, 23% gave no information at all about what was done to optimize the hits. Glycerol was used as a cryoprotectant in half the papers, and was used equally often at either 11–20 or 21–30%. Over 80% of the papers gave no indication of how reliable the diffraction was for the crystals.
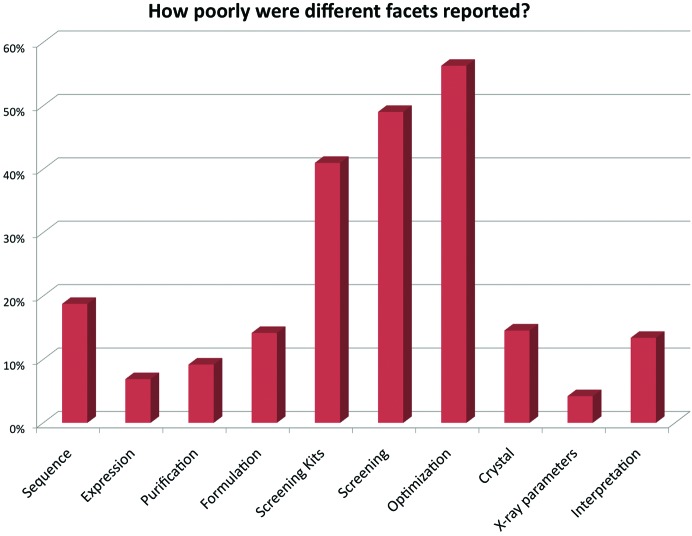
The last few questions of the survey asked about how well the paper did in capturing information about different parts of a crystallization experiment. According to the (necessarily subjective) surveyors, over 60% of the papers did not contain all the information necessary to replicate the experiment, with the most commonly cited lack being details about the optimization process. Fig. 1 ▶ shows more details of what these surveyors believed was poorly described in the papers.
Figure 1.
Each surveyor was asked to comment on whether or not more information was needed for a number of different areas in the paper. The graph shows the percentage of papers that the surveyors thought needed more detail in that part of the paper. Overall, the optimization of the initial hits obtained from screening was the most poorly described, with almost 60% of the papers surveyed providing no or limited information about the process. The x-axis labels refer to the following points: What part of the experimental description needs more detail? (click all that apply); Description of the molecule under study (e.g. no precise sequence provided); Description of the expression of the molecule; Description of the purification of the molecule; Description of the formulation of the molecule (what buffer is it in when crystallized, at what concentration?); Description of the screening conditions (which screens); Description of the screening setup (drop volumes, ratios, temperatures, robotic or manual setup); Description of the optimization (what was varied); Description of the optimized crystal (size, morphology); Crystallographic parameters (unit cell, diffraction limit); It’s all sort of there, but one has to do a lot of interpretation to get all the details.
3.3. Crystallization chemicals
The online survey made the assumption that each crystallization communication would have only one final condition associated with it. This was overcome on the fly by submitting a new survey for each crystallization condition, thus there were 320 completed surveys for 264 papers. A summary of the conditions used in the papers is given in Supplementary Tables 1 and 2. The chemicals used follow the same trends as reported previously (Peat et al., 2005 ▶): namely seven of the top 25 chemicals are polyethylene glycols (PEGs), with PEG 3350 being the most popular. The most popular buffers are Tris and HEPES, following the trend in pH where there is a peak at pH 7 to 8 for crystallization. Ammonium sulfate continues to be a popular crystallizing agent; citrate and acetate are the next most popular compounds being both precipitating agents and buffers; and rounding out the top ten also gives us magnesium chloride and the buffer MES. Interestingly, sodium malonate, which was presented as a useful crystallization salt in 2001 (McPherson, 2001 ▶), was only found once in the successful conditions.
3.4. Where do these papers originate?
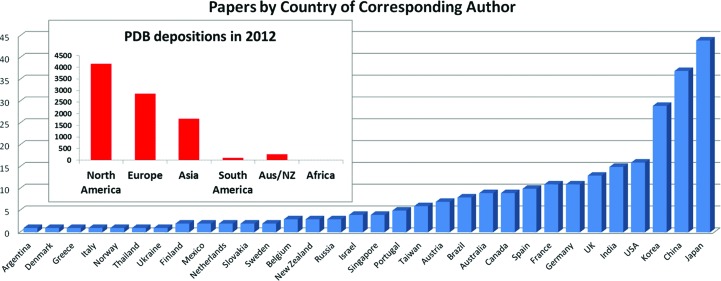
There was a clear geographical bias of the papers. Fig. 2 ▶ shows the country associated with the corresponding author for the crystallization communications published in 2012. Of the 32 different countries contributing crystallization communications in 2012, three countries, Korea, China and Japan, between them contributed over 40% of the papers. The same figure shows the distribution of the origin of the structures deposited during 2012, from the wwPDB website. The majority (over 40%) of the depositions come from the USA.
Figure 2.
The count of papers from each country (according to the e-mail address of the corresponding author). There were only 16 papers in 2012 from the USA, compared to 44 from Japan. Assuming that Japan and US have proportionally the same number of researchers per capita, then normalizing for population (estimated 128 million for Japan, 314 million for the USA) Japan published almost 10× the number of crystallization papers as the USA. The insert shows data from the wwPDB (http://www.wwpdb.org/stats.html) on the source of the PDB depositions for the year 2012.
3.5. Structures associated with the papers
The search tools from the PDB were used to try to map the work described in the crystallization communications in 2012 to PDB codes. The mapping was done using molecule name and author name, and verifying this by checking cell dimensions and space group. The date of the PDB deposition had to be 2011 or later. Surprisingly, most of the Acta F crystallization communications do not map to PDB codes at all. Table 2 ▶ enumerates the list of Acta F communications for which PDB codes could be found by searching the PDB for author, molecule name, cell dimensions and space group (the search was carried out in May 2013). At that time, only about 100 of the PDB codes (∼1%) of the deposited structures in 2013 mapped to an Acta F paper.
Table 2. A list of 74 crystallization reports, out of the 264 published in 2012, where one (or more) PDB deposition code(s) could be found.
The papers are identified by the doi. Five more PDB codes (4bej, 4fqn, 4eqi, 4asn, 4dcf) mapped to a molecule described in a crystallization communication, but not the authors; this might reflect a situation where the authors of the paper were scooped on the final structure by a competing group.
| Paper doi | Code 1 | Code 2 | Code 3 | Code 4 |
|---|---|---|---|---|
| 10.1107/S1744309112038705 | 3vsg | 3vsh | 3vsj | 3vsi |
| 10.1107/S1744309112031181 | 4bc2 | 4bc3 | 4bc4 | 4bc5 |
| 10.1107/S1744309111048111 | 3pqd | 3pqf | 3pqe | |
| 10.1107/S1744309112001327 | 3vki | 3vjp | 3tee | |
| 10.1107/S1744309111048743 | 4ad8 | 4abx | 4aby | |
| 10.1107/S1744309112045551 | 4ei7 | 4ei8 | 4ei9 | |
| 10.1107/S1744309112039036 | 1yiv | 4a8z | 4a1y | |
| 10.1107/S1744309112035294 | 4ae2 | 4aej | 4ak3 | |
| 10.1107/S1744309112038936 | 4gel | 4gem | 4gen | |
| 10.1107/S1744309112032447 | 3i98 | 3q3l | ||
| 10.1107/S1744309111047634 | 3t35 | 3t34 | ||
| 10.1107/S1744309111047920 | 3thc | 3thd | ||
| 10.1107/S1744309111049530 | 4aaz | 4ab0 | ||
| 10.1107/S1744309111046318 | 4eq2 | 4eq3 | ||
| 10.1107/S1744309112000590 | 4f3h | 4f48 | ||
| 10.1107/S1744309112004940 | 2yfa | 2yfb | ||
| 10.1107/S1744309112004691 | 3vpe | 3vqz | ||
| 10.1107/S1744309111054212 | 4a0t | 4a0u | ||
| 10.1107/S1744309112017307 | 4axc | 4acd | ||
| 10.1107/S1744309112028229 | 4f33 | 4f3f | ||
| 10.1107/S1744309112025651 | 4fbm | 4fbl | ||
| 10.1107/S1744309112033088 | 4iu2 | 4iu3 | ||
| 10.1107/S1744309112000474 | 3sv0 | |||
| 10.1107/S1744309112004952 | 4e6z | |||
| 10.1107/S1744309112032721 | 1h5o | |||
| 10.1107/S1744309112033052 | 2hmc | |||
| 10.1107/S1744309112020301 | 3q6i | |||
| 10.1107/S1744309111049116 | 3qe6 | |||
| 10.1107/S1744309111048688 | 3t37 | |||
| 10.1107/S1744309112018672 | 3ucf | |||
| 10.1107/S1744309107005441 | 3uk7 | |||
| 10.1107/S1744309111050597 | 3uk7 | |||
| 10.1107/S1744309112003740 | 3vgi | |||
| 10.1107/S1744309112025328 | 3vsu | |||
| 10.1107/S1744309112032435 | 3vu7 | |||
| 10.1107/S1744309112016004 | 3w3s | |||
| 10.1107/S1744309112011888 | 3znu | |||
| 10.1107/S1744309112008445 | 4a5u | |||
| 10.1107/S1744309111055187 | 4abx | |||
| 10.1107/S1744309112009736 | 4akl | |||
| 10.1107/S1744309112020180 | 4am1 | |||
| 10.1107/S174430911203895X | 4aya | |||
| 10.1107/S1744309111056028 | 4b7o | |||
| 10.1107/S1744309112031077 | 4den | |||
| 10.1107/S1744309112030874 | 4dkx | |||
| 10.1107/S1744309111052109 | 4dmz | |||
| 10.1107/S1744309111049529 | 4doh | |||
| 10.1107/S1744309111055928 | 4e0v | |||
| 10.1107/S1744309112015692 | 4e1o | |||
| 10.1107/S1744309112041401 | 4e1v | |||
| 10.1107/S174430911201740X | 4e4h | |||
| 10.1107/S1744309112006070 | 4e6s | |||
| 10.1107/S1744309112022208 | 4emo | |||
| 10.1107/S1744309111050913 | 4eq5 | |||
| 10.1107/S1744309112044417 | 4eqv | |||
| 10.1107/S1744309112016569 | 4es7 | |||
| 10.1107/S1744309112020313 | 4es8 | |||
| 10.1107/S1744309112044041 | 4exw | |||
| 10.1107/S1744309112022075 | 4f9j | |||
| 10.1107/S174430911201665X | 4fdf | |||
| 10.1107/S1744309112019082 | 4g0b | |||
| 10.1107/S1744309112045563 | 4g0r | |||
| 10.1107/S1744309112024372 | 4g3l | |||
| 10.1107/S1744309112027236 | 4gjt | |||
| 10.1107/S1744309112019306 | 4gtw | |||
| 10.1107/S1744309112045447 | 4h2f | |||
| 10.1107/S1744309112000838 | 4h7m | |||
| 10.1107/S1744309112047070 | 4hac | |||
| 10.1107/S1744309112042212 | 4htg | |||
| 10.1107/S1744309112040638 | 4i9a | |||
| 10.1107/S1744309112023603 | 4ic5 | |||
| 10.1107/S1744309112025213 | 4ij5 | |||
| 10.1107/S1744309112028989 | 4iq8 | |||
| 10.1107/S1744309112032526 | 4ix9 |
4. Discussion
One of the more surprising things to come out of this analysis was how few of the Acta F reports seemed to map to PDB codes. Using the search tools provided in the PDB, we estimate that just over a quarter of the 264 papers surveyed had one or more associated PDB code(s) by May 2013. Necessarily, all of the crystal systems described in the communications have significant diffraction associated with them. Are three quarters of the projects awaiting phase information? Or are they yet to be deposited? In at least five cases, there was a recent deposition for a similar molecule found in the PDB, deposited by people other than the authors of the crystallization paper. It could be that the crystallization communication was a rescue strategy for the team that was beaten to the structure.
Given that crystallization is rarely as simple as setting up a commercial screen and flash-cooling the crystals that grew in it using a cryosolution made by adding 20% glycerol to the well solution, we wonder why more groups don’t make more of the opportunity provided by these specialist crystallization papers to communicate how they solved their expression, purification and crystallization problems. There can be incredible amounts of finesse in getting to a well diffracting and appropriate crystal; these are the stories that inspire and educate us all. Yet the majority of papers don’t seem to discuss the details of the process at all. It was disturbing that even early career PhD students generally did not find any useful/novel techniques or tips that they wanted to try themselves after reading through 20 or more of these papers.
Of the two most likely ways that a crystallization paper could be useful, replication of a published condition does not require any information about the initial screening, whereas information about the robustness of the process (e.g. how reliably the crystals grew and how reliably they diffracted) is of great interest. The collective information about screening strategies is highly pertinent to those starting a new crystallization project; here the information about the robustness of any given system is less relevant. Table 3 ▶ gives a comparison of what we believe is the information needed in both cases.
Table 3. Starting a new crystallization project requires different information from trying to replicate a published crystallization condition.
Generally replication of previously published work is to allow for more systematic studies on the system, such as small-molecule binding studies. In this case, information about the ease of obtaining, growing and handing the crystals is paramount. For investigators starting a new project, the relevant information would be what are the most successful commercial screens, and how many drops are required for a ‘normal’ screening and optimization. Ideally, the mandatory information required to describe a crystallization experiment should be described very clearly, for example as has been done for SAX data (Jacques et al., 2012 ▶).
| Replication | New project guide |
|---|---|
| Construct details | What tags are used |
| Expression system | Cleave off tag before crystallization |
| Purification steps | How many screening drops to try |
| Yield of protein | What drop sizes/ratios to try |
| Can the protein be frozen? | What experimental setups to use |
| Additives/treatment required for crystallization | What screens to use |
| Optimized crystallization condition | How long to wait for a hit |
| Range of the optimized condition | What a starting hit looks like |
| Reliability of crystal growth | What optimization strategies work |
| Robustness of crystals | How much optimization to attempt |
| Reliability of good diffraction |
If one has the goal of reproducing a crystal, then any source of information about the specific crystallization protocol for that target is a starting point: the REMARK 280 from a PDB deposition; a materials and methods section of a structure paper or a dedicated crystallization paper. In this case the dedicated crystallization paper should make the process easier by describing possible pitfalls. However, anyone who has reproduced a crystal described previously in the literature can attest to how much tweaking is invariably required, and that the information about the original conditions, whatever the source, is more of an affirmation that crystals can be made, rather than a sure-fire recipe of how to make them.
The collection of crystallization data has historically been very limited; most public databases capture only the final, successful crystallization condition, and even those data are sparse (Newman et al., 2012 ▶). The crystallization community is in the somewhat bizarre situation of being better at growing crystals than at knowing how to grow crystals. It would be of great benefit to be able to point to a data resource that would enable one to tackle questions like ‘What screen(s) are others finding useful (and I should use)?’, ‘How many drops should I set up?’ and even ‘What is the most appropriate protein concentration range for these experiments?’.
The collection and collation of these types of collective crystallization information is potentially the real strength of specialist crystallization communications. Unfortunately, as is shown in Fig. 1 ▶, the details about the actual crystallization experiments performed (which screens were used; how the screening was done; and how any optimization was done) are the least well described of the experimental aspects of the crystallization papers. This could be a result of these being more difficult to capture than details about the expression (which was generally ranked as being well reported), but could be a result of a belief that only the final, optimized condition needs to be reported. However, there were at least two papers where even the final, optimized condition was not reported.
One of the ways to ameliorate this paucity of detail would be to encourage the wider use of templates for crystallization communications. The IUCr already has a template which can be used; see http://publbio.iucr.org/publbio. The use of templates is helpful in that it gives the authors clear guidelines as to what is expected of these types of papers. One of the strengths of templates is that they can be built to include current standards, for example in chemical naming, so that the papers built using the template would have consistent names and format styles for chemicals, and the description of crystallization. Other features would be menus prepopulated with information, for example vector names or crystallization screens, to help ensure accuracy and consistency. Ideally, a data standard (possibly based on mmCIF format) could be used store the final crystallization conditions and this same representation could be easily transferred to data repositories like the PDB. By making this easy, we would start to move towards standards in naming and format in crystallization which now simply don’t exist, and which are absolutely required for future data-mining projects. The survey questions used here might be a good start for setting the requirements of the template, although what the community finds important will be an ongoing discussion.
It was interesting that almost 75% of the papers surveyed described the crystallization of a single, soluble protein sample. Just a quick look at the structure papers being published in the more prominent journals suggests that structure papers need to report membrane protein structures and/or the structures of protein complexes in order to be published, so that the crystallization papers are not representative of current practices suitable to produce the ‘hot’ structures. The crystallization communications have no timeframe associated with them, neither when the project was initiated, nor how long it continued. Is the bias towards Crystal Screen and Crystal Screen II from Hampton Research a reflection of the abiding appropriateness of these screens, or is it a reflection that these were amongst the first screens available commercially? Is the lack of sodium malonate in the list of successful conditions due to this being older work? Similarly, papers which use 96-well plates will necessarily be describing work performed in the last 15 years (as these plates became available in the latter part of the last century) but drops set up in 24-well plates may well be recent work set up in a laboratory without access to automation.
Why aren’t researchers from the USA (for example) publishing more crystallization reports? If the metrics for academic success are simply based on paper number, then publishing crystallization reports would have obvious benefits to the researcher. However, for success metrics that involve non-self-citation rates and the impact factor of the journal as well, publication of minor papers in low-impact-factor journals can be quite deleterious. In this case it would be better to have one paper with supplementary information in a high-impact journal. The information routinely captured in supplementary information is the bare minimum of the information required for replication, and is very unlikely to contain the information likely to help develop and refine crystallization strategies. Furthermore, supplementary information is rarely rigorously reviewed, and any references cited in the supplementary material do not get added to the citation counts for those references. Even if supplementary documentation did contain the information useful for developing snapshots of the current best practices, it is not clear how that data could be collated, and the nature of this information requires it to be collective to be useful.
What is the goal of the current crystallization communication? Is it to provide a training tool for young scientists? Is it to capture crystallization information about a specific system? Has it become a consolation prize for when something prevents a ‘better’ paper? We believe that there needs to be a clearer mandate as to the reason for these papers.
Currently, around 90% of crystallization communications that are submitted are eventually published. Irrespective of what the final mandate of these papers turns out to be, an author of a crystallization paper would like their work to be interesting and useful to their peers. Below we suggest some approaches which might increase the impact of crystallization papers, and may encourage a greater understanding of the trials that each of us face in the world of macromolecular crystallization.
(1) Instigate a policy by which only the scientists who contributed to the ‘hands-on’ laboratory work described are listed as authors on the paper; the research director or laboratory head would be acknowledged formally within the paper rather than by authorship. The wet-work is most likely to be done by early career scientists and technical personnel and both groups would gain from having (and writing) extra publications, even if not high impact.
(2) Encourage the greater use of formal templates, which have been set up to include selection menus to ensure that crystallization data are reported in a standard format, using consistent and unambiguous names.
(3) Have a two-stage process towards publication of crystallization communications. The first, required step would be to fill in a template, essentially answering the questions asked in the survey that was used for this analysis, or similar questions. Only after that information has been provided would the manuscript be passed through the editorial process to see if it contains information that would intrigue or educate the crystallization community. The first step means that the statistics could be captured about what screens are being used and other such ‘collectively useful’ information, without overloading the crystallization community with papers that contain no novelty.
(4) Change the policy of publishing crystallization communications. To enable the authors to ‘hold’ publication until the associated structure paper is published would allow crystallization communications to smoothly take over the role currently played by the supplementary information, and would allow this information to be more widely disseminated and used for overview analyses. This is analogous to the ‘hold’ concept for coordinates deposited in the PDB.
5. Conclusions
The crystallization communications published by Acta Crystallographica Section F in 2012 give a limited snapshot of the techniques and chemicals being used to crystallize single proteins. Unfortunately, they offer little insight towards understanding the crystallization of complexes of biomacromolecules, nor do they provide much information regarding integral membrane protein crystallization. In order to capture and push our limited knowledge of this complex field further, we need to encourage the publication of crystallization reports by the groups doing these types of cutting-edge structures. We believe that there is great potential for the Acta F crystallization communications to be a fascinating and useful part of the literature in structural biology, but this will require some changes. We suggest a number of possible routes for increasing the impact of these communications: firstly, to encourage the more junior scientists and technical staff to take a leading role in the publication of these reports; secondly, to require the use of a standard template for these papers; thirdly, to implement a requirement for the crystallization information within a paper before the paper is considered for publication, and finally, to have a ‘hold’ system for crystallization reports that allows simultaneous publication with the structure report and/or PDB deposition.
Supplementary Material
Raw data from surveying papers.. DOI: 10.1107/S1744309113014152/wd5209sup1.xlsx
Consistency analysis, raw data.. DOI: 10.1107/S1744309113014152/wd5209sup2.xlsx
Supplementary material file. DOI: 10.1107/S1744309113014152/wd5209sup3.pdf
Acknowledgments
We thank Louise Jones and Peter Strickland for access to the papers from 2012, and Manfred Weiss and Howard Einspahr for helpful and animated discussions.
References
- Begley, C. G. & Ellis, L. M. (2012). Nature (London), 483, 531–533. [DOI] [PubMed]
- Bornmann, L., Mutz, R. & Daniel, H. (2008). J. Am. Soc. Inf. Sci. Technol. 59, 830–837.
- Boutet, S. et al. (2012). Science, 337, 362–364.
- Chapman, H. N. et al. (2011). Nature (London), 470, 73–77.
- Ferré-D’Amaré, A. R. & Burley, S. K. (1997). Methods Enzymol. 276, 157–166. [DOI] [PubMed]
- Jacques, D. A., Guss, J. M., Svergun, D. I. & Trewhella, J. (2012). Acta Cryst. D68, 620–626. [DOI] [PubMed]
- McPherson, A. (2001). Protein Sci. 10, 418–422. [DOI] [PMC free article] [PubMed]
- Newman, J., Bolton, E. E., Müller-Dieckmann, J., Fazio, V. J., Gallagher, D. T., Lovell, D., Luft, J. R., Peat, T. S., Ratcliffe, D., Sayle, R. A., Snell, E. H., Taylor, K., Vallotton, P., Velanker, S. & von Delft, F. (2012). Acta Cryst. F68, 253–258. [DOI] [PMC free article] [PubMed]
- Newman, J., Fazio, V. J., Lawson, B. & Peat, T. S. (2010). Cryst. Growth Des. 10, 2785–2792.
- Peat, T. S., Christopher, J. A. & Newman, J. (2005). Acta Cryst. D61, 1662–1669. [DOI] [PubMed]
- Seeber, F. (2008). Nature (London), 451, 887. [DOI] [PubMed]
- Weiss, M. S., Einspahr, H., Baker, E. N., Dauter, Z., Kaysser-Pyzalla, A., Kostorz, G. & Larsen, S. (2010). J. Appl. Cryst. 43, 1285–1286.
- Wilson, W. W. (2003). J. Struct. Biol. 142, 56–65. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw data from surveying papers.. DOI: 10.1107/S1744309113014152/wd5209sup1.xlsx
Consistency analysis, raw data.. DOI: 10.1107/S1744309113014152/wd5209sup2.xlsx
Supplementary material file. DOI: 10.1107/S1744309113014152/wd5209sup3.pdf