Abstract
Background
The influence of variants at the 9p21 locus on melanoma risk has been reported through investigation of CDKN2A variants through candidate gene approach as well as by genome wide association studies (GWAS).
Methods
In the present study we genotyped, 25 SNPs that tag 273 variants on chromosome 9p21 in 837 melanoma cases and 1154 controls from Spain. Ten SNPs were selected based on previous associations, reported in GWAS, with either melanocytic nevi or melanoma risk or both. The other 15 SNPs were selected to fine map the CDKN2A gene region.
Results
All the 10 variants selected from the GWAS showed statistically significant association with melanoma risk. Statistically significant association with melanoma risk was also observed for the carriers of the variant T-allele of rs3088440 (540 C>T) at the 3’ UTR of CDKN2A gene with an OR 1.52 (95% CI 1.14-2.04). Interaction analysis between risk associated polymorphisms and previously genotyped MC1R variants, in the present study, did not show any statistically significant association. Statistical significant association was observed for the interaction between phototypes and the rs10811629 (located in intron 5 of MTAP). The strongest association was observed between the homozygous carrier of the A–allele and phototype II with an OR of 15.93 (95% CI 5.34-47.54).
Conclusions
Our data confirmed the association of different variants at chromosome 9p21 with melanoma risk and we also found an association of a variant with skin phototypes.
Background
Cutaneous malignant melanoma is one of the cancer types with rapid increase in incidence rates in many Caucasian populations worldwide [1,2]. The risk factors associated with melanoma include environmental sun exposure and genetically determined host factors as skin color, eye color, hair color, freckling and the presence and number of nevi [2-6]. One of the most significant risk factors in melanoma is the family history of the disease. Approximately, 10% of melanoma is associated with familial predisposition with affected first or second-degree relatives. Out of those 20%-40% of the familial melanoma is linked to chromosome 9p21 locus and a proportion of 9p21 linked families carry disease-segregating germline mutations in cyclin-dependent kinase inhibitor 2A (CDKN2A) gene [7]. Over the years several genome wide association studies (GWAS) have identified a number of low penetrance variants at the locus 9p21 that were associated with risk of either melanoma or cutaneous nevi or both [8-12]. The locus has also been previously investigated through candidate gene approach with particular emphasis on the CDKN2A gene for detection of variants associated with risk of melanoma [13-16].
Two GWAS showed the association of a total of 10 single nucleotide polymorphisms (SNPs) at 9p21 locus with either cutaneous nevi or melanoma risk or both. The polymorphisms rs751173, rs1341866 (both intergenic) and rs10811629 located in intron 5 of methylthioadenosine phosphorylase (MTAP) have been associated with cutaneous nevi while the three intergenic polymorphisms rs4636294, rs2218220, rs1335510 and the two intronic rs10757257 and rs7023329 with cutaneous nevi and melanoma [8,9]. Another study showed association of the imputed intergenic variant rs935053 with melanoma risk. Moreover, stepwise regression analysis of 11 tag SNPs covering a 2 Mb region at the 9p21 locus showed the association between the polymorphism rs1011970 and melanoma [9]. All the polymorphisms except rs1011970 were located in a 132.9 Kb region containing the methylthioadenosine phosphorylase (MTAP) gene. The polymorphism rs1011970 was located in antisense non coding RNA (ANRIL) gene. The association of the 9p21 locus with melanoma risk was replicated in subsequent GWAS and in a recent meta-analysis [10-12,17].
The association of the polymorphisms 540 C>T (rs3088440) and 500 C>G (rs11515) at the 3’ untranslated region of CDKN2A with melanoma risk investigated by candidate gene approach has remained ambiguous. An earlier study based on familial melanoma in an Australian population showed an association of the variant 500 C>G with risk of melanoma [13]. Another study based on sporadic primary melanomas showed statistically significant association of the polymorphism 540 C>T with melanomas but no statistically significant association was reported for the polymorphism 500 C>G. The study showed the linkage disequilibrium of the variant 500 C>G with a polymorphism in intron 1 of the adjacent cyclin-dependent kinase inhibitor 2B (CDKN2B) gene [14]. However, later studies could not replicate the association of the two polymorphisms with melanoma risk [15,16].
In the present study, we genotyped 25 polymorphisms at 9p21 locus, selected by the use of tagging approach and on the basis of previous reported associated variants in GWAS, in Spanish melanoma cases and matched controls. We confirmed the association between melanoma risk and ten polymorphisms that were previously shown in GWAS to be associated with nevi or melanoma risk or both. Of the other 15 variants selected through tagging SNP approach to fine map CDKN2A gene we found an association between the rs3088440 variant and melanoma risk.
Methods
Study populations
The present study included 837 Spanish cutaneous melanoma patients. The cutaneous melanoma patients were recruited at the Department of Dermatology, Instituto Valenciano de Oncologia, a referral skin cancer center for the provinces of Valencia, Alicante and Castellón, with ~5 million population. Blood samples from melanoma patients with histopathologically confirmed diagnosis were collected between 2000 and 2007. Clinical and pathological data from the patients were prospectively collected since January 2000 through the review of medical history, personal interviews and clinical examination by expert dermatologists. The patients were followed-up for a median of 88.8 months (95% CI 83.1-94.4). The tumors were classified according to the AJCC stage system for pathological stage and the relative Breslow thickness was reported. The phenotypic characteristics of skin in cases were classified as Fitzpatrick phototypes after examination by a trained dermatologist. Information about hair and eye color was also documented. Spanish control included 1154 ethnically matched disease-free controls-individuals. Disease-free and ethnically matched healthy control subjects were recruited at the Transfusion Center of Valencia, Spain. The phenotypic characteristics of skin in controls were classified as Fitzpatrick phototypes based on a self-reported questionnaire.
All the participants in the study signed an informed consent and the study protocol was approved by institutional ethic board of Instituto Valenciano de Oncologia.
Selection and genotyping of the polymorphisms
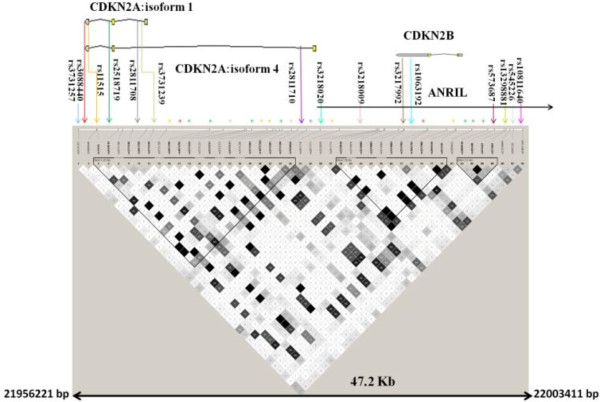
The polymorphisms on chromosome 9p21 were selected according to two criteria: polymorphisms previously reported to be associated in published GWAS on melanoma and nevi and polymorphisms selected through tagging approach to fine map the CDKN2A gene. Ten polymorphisms reported to be associated with melanoma and cutaneous nevi or both were included in the study (Figure 1).
Figure 1.
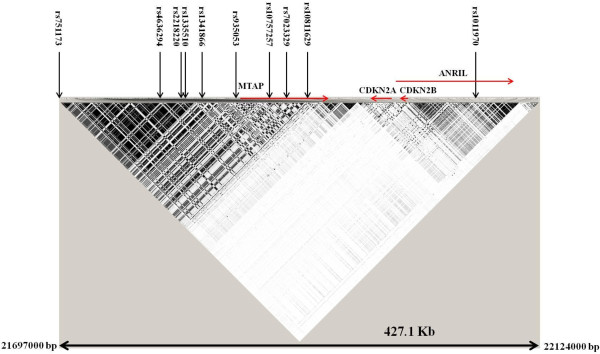
Linkage disequilibrium map of chromosome 9p21 from 21697000 bp to 22124000 bp. The linkage disequilibrium map derived from HapMap (release #27) shows the SNPs associated with cutaneous nevi or melanoma risk or both in GWAS.
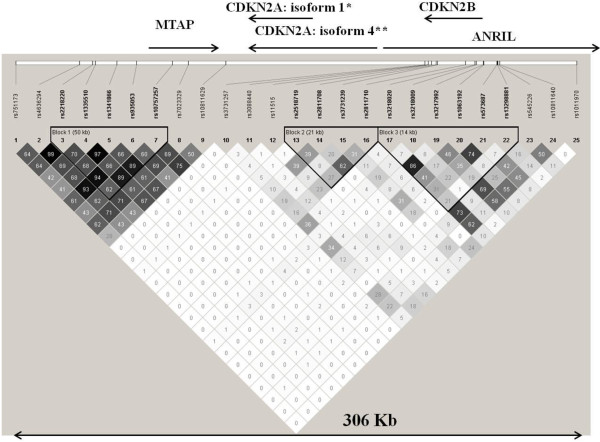
The selection of the polymorphisms in order to fine map the CDKN2A gene was done using data on the Caucasian population from HapMap database (release #27). The 47.2 Kb (chromosome 9: 21956221–22003411; build 36.3) region encompassing CDKN2A and the nearby CDKN2B was scanned and 12 polymorphisms with a minor allele frequency ≥0.10 were identified using tagger program in Haploview using a minimum r2 of 0.8 (Figure 2). A total of 25 polymorphisms were selected on chromosome 9p21 and genotyped in the Spanish melanoma cases and controls (Additional file 1: Table S1).
Figure 2.
Linkage disequilibrium map of chromosome 9p21 from 21956221 bp to 22003411 bp. The map derived from HapMap (release #27) reports the genotyped tagging SNPs.
Genotyping of the selected polymorphisms was carried out using a PCR based allelic discrimination method (KBiosciences UK). The amplification was carried out in a final volume of 4 μl using 5 ng of DNA, 2 μl 2X reaction mix (KBiosciences, UK), 0.06 μl of the solution containing 100 nM of two forward primers specific for two alleles, and one reverse primer mix. The reaction was performed under thermocycling conditions with an initial denaturation at 94°C for 15 minutes, followed by 36–46 cycles of 20s at 94°C and 60s of annealing (temperatures ranging from 60-50°C). Genotypes in amplified products were determined by differences in VIC and FAM fluorescent level in plate read operation on ABI PRISM 7900HT (Applied Biosystems, Foster City, CA) using SDS 1.2 Software. Post operation data were transferred as Microsoft Excel files and converted into genotype informations.
The samples for which genotyping failed in Kaspar assay were randomly sequenced. PCR for sequencing was carried out in a total volume of 10 μl. The reaction contained 10 ng of DNA template, 1.0 μl of 10X reaction buffer, 0.11 mM of each dNTP, 0.20 μM of each primer (Additional file 2: Table S2), 1.50 mM of MgCl2 and 0.2 U (TAQ POL) of DNA Taq Polymerase (Genaxxon BioScience GmbH, Germany). Thermocycling conditions used were 94 C for 5 min, followed by 3 cycles of 94°C for 45 seconds, annealing temperature (Additional file 2: Table S2) for 45 seconds, 72°C for 45 seconds followed by additional 35 repeated cycles of 94°C for 30 seconds, annealing temperature (Additional file 2: Table S2) for 30 seconds and 72 C for 45 sec. The final step was 72°C for 5 minutes.
The amplified fragments were checked on 1% agarose gel. PCR product were purified by incubation for 30 minutes at 37 C with 0.75 μl of ExoSAP-IT®(USB Corporation, Cleaveland, OH), followed by an incubation for 15 minutes at 85°C. Cycle sequencing reaction on purified PCR products were performed using specific primers (Additional file 2: Table S2). The sequencing reaction was performed using Big Dye® Terminator Cycle Kit (Applied Biosystem, Foster City, CA). The reaction products were precipitated with isopropanol, washed in 70% ethanol and the precipitate was suspended in 25 μl of water. The 96 well plates containing DNA were loaded on an ABI prism 3100 Genetic analyzer and the data were analyzed using a sequencing program (Applied Biosystem).
Statistical analysis
Genotype frequencies of all SNPs were tested in control subjects for deviation from Hardy-Weinberg equilibrium using Pearson’s X2 test. For association study, 63 patients with melanoma in situ (AJCC stage 0) were excluded from data analysis. The association between cutaneous melanoma and genotype of the polymorphisms was evaluated by odds ratios (OR), 95% confidence intervals (CI) and P values, using logistic regression (SAS version 9.2, SAS institute Inc., NC). Logistic regression models included age and sex as covariates. Analysis was also performed using a model selection that included all covariates.
Gene-gene and gene-host factor interactions were carried out after a model selection based on likelihood ratio tests (LTR). The interaction analysis between the variants that showed association with the risk of melanoma and previously genotyped melanocortin receptor 1 (MC1R) variants was carried out by categorizing both cases and controls into carriers and non-carriers [18]. For MC1R categories selected were carriers of any (V60L, D84E, V92M, R151C, I155T, R160W, R163Q and D294H) MC1R variants versus non-carriers and analysis was also carried out by grouping red hair color (RHC; D84E, R151C, R160W and D294H) and non-RHC variants separately. All statistical tests were performed in SAS 9.2 software.
Interaction contrast ratio ICR to test departure from additivity (> or < 0) and the multiplicative interaction index MII to test departure from multiplicativity (> or < 1) were also calculated in SAS 9.2. Confidence intervals and the p-values were calculated using bootstrap methods with 10000 simulations (SAS 9.2).
Bioinformatic analysis
The estimate of linkage disequilibrium between the polymorphisms in Spanish cases and controls combined was performed using SNP_tool package. The converted data were processed in the Haploview software (version 4.2) for linkage disequilibrium estimation.
Analysis of expression quantitative trait loci (eQTL) in the eQTL browser (eqtl.uchicago.edu/Home.html) was performed for the statistically significant associated polymorphisms with melanoma risk and the tagged variants.
Results
The present study included 837 cutaneous melanoma cases from Spain and 1154 disease-free ethnically matched controls. The cases comprised of 459 women and 378 men with mean and median age of 51.5 (±16.1) and 53 (inter-quartile range: 39–65) years, respectively. Spanish controls were composed of 478 women and 676 men with mean and median age of 45 (±7.3) and 44 (inter-quartile range: 39–50) years, respectively. In cases and controls phenotypic traits like Fitzpatrick phototype, hair and eye color were associated with risk of melanoma in an expected manner as previously reported [19]. The cases and controls were genotyped for a total of 25 polymorphisms that tagged a total of 273 variants at the 9p21 locus. Ten SNPs were selected based on GWAS and an additional 15 were selected to fine map CDKN2A gene. Sixty-three melanoma patients with AJCC stage 0 were excluded from data analysis. Information on characteristics of melanoma patients and controls subjects are reported in our previous study [19]. The genotype frequencies for all the variants included in the study were, in controls, in accordance with Hardy-Weinberg equilibrium.
The data analysis showed that of the 25 genotyped variants, all 10 variants selected on the basis of GWAS showed statistically significant association with melanoma risk. Of the 15 variants selected to fine map CDKN2A gene, the rs3088440 (3’UTR CDKN2A) variant, showed statistically significant association with melanoma risk. In addition the variant T-allele of the tagging rs2811710 variant also showed statistically significant association with the risk of melanoma. None of the remaining 13 tagging polymorphisms showed statistically significant association with the disease risk.
The rs751173 polymorphism located in an intergenic region between the interferon, epsilon (IFNE) and the MTAP genes showed association with a statistical significant increased melanoma risk (Figure 1). The carriers of the variant C-allele were at an increased risk of developing melanoma (OR 1.25, 95% CI 1.01-1.55) (Table 1). The rs751173 polymorphism tagged (with r2 ≥0.8) 22 SNPs according to HapMap database. All tagged SNPs were located in the intergenic region between IFNE and MTAP genes.
Table 1.
Analysis of genotype data from melanoma cases and controls for polymorphisms on chromosome 9p21 locus
| SNP | Cases (%) | Controls (%) | OR | 95% CI | P value |
|---|---|---|---|---|---|
|
rs751173 |
|
|
|
|
|
| TT |
211 (27.2) |
358 (31.1) |
Reference |
|
|
| TC |
384 (49.6) |
573 (49.7) |
1.18 |
0.94 –1.47 |
|
| CC |
179 (23.1) |
221 (19.2) |
1.43 |
1.09-1.87 |
0.04 |
| TC+CC |
563 (72.7) |
794 (68.9) |
1.25 |
1.01-1.54 |
0.04 |
| T allele |
806 (52.1) |
1286 (56) |
Reference |
|
|
| C allele |
742 (47.9) |
1015 (44.1) |
1.19 |
1.04–1.37 |
0.01 |
| MAF |
0.48 |
0.44 |
|
|
|
|
rs4636294 |
|
|
|
|
|
| GG |
169 (21.9) |
280 (24.3) |
Reference |
|
|
| AG |
375 (48.5) |
605 (52.5) |
1.07 |
0.84–1.36 |
|
| AA |
229 (29.6) |
268 (23.2) |
1.41 |
1.08–1.85 |
0.02 |
| AG+AA |
604 (78.1) |
873 (75.7) |
1.17 |
0.93–1.47 |
0.16 |
| G allele |
713 (46.1) |
1165 (50.5) |
Reference |
|
|
| A allele |
833 (53.9) |
1141 (49.5) |
1.19 |
1.04–1.36 |
0.01 |
| MAF |
0.54 |
0.50 |
|
|
|
|
rs2218220 |
|
|
|
|
|
| TT |
167 (21.6) |
281 (24.4) |
Reference |
|
|
| CT |
379 (49) |
607 (52.7) |
1.09 |
0.86–1.39 |
|
|
CC |
228 (29.5) |
264 (22.9) |
1.45 |
1.11–1.91 |
0.01 |
| TC+CC |
607 (78.4) |
871 (75.6) |
1.20 |
0.96–1.51 |
0.12 |
| T allele |
713 (46.1) |
1169 (50.7) |
Reference |
|
|
| C allele |
835 (53.9) |
1135 (49.3) |
1.21 |
1.05–1.38 |
0.007 |
| MAF |
0.54 |
0.49 |
|
|
|
|
rs1335510 |
|
|
|
|
|
| TT |
323 (41.8) |
406 (35.4) |
Reference |
|
|
| TG |
352 (45.6) |
568 (49.5) |
0.79 |
0.64–0.97 |
|
| GG |
97 (12.5) |
173 (15.1) |
0.67 |
0.49–0.91 |
0.01 |
| TG+GG |
449 (58.2) |
741 (64.6) |
0.76 |
0.63–0.93 |
0.007 |
| T allele |
998 (64.6) |
1380 (60.2) |
Reference |
|
|
| G allele |
546 (35.4) |
914 (39.8) |
0.81 |
0.71–0.94 |
0.004 |
| MAF |
0.35 |
0.40 |
|
|
|
|
rs1341866 |
|
|
|
|
|
| TT |
316 (40.8) |
398 (34.6) |
Reference |
|
|
| TC |
361 (46.6) |
576 (50) |
0.81 |
0.66–0.99 |
|
| CC |
97 (12.5) |
177 (15.4) |
0.66 |
0.49–0.90 |
0.02 |
| TC+CC |
458 (59.2) |
753 (65.4) |
0.77 |
0.63–0.94 |
0.01 |
| T allele |
993 (64.2) |
1372 (59.6) |
Reference |
|
|
| C allele |
555 (35.9) |
930 (40.4) |
0.82 |
0.71–0.94 |
0.004 |
| MAF |
0.36 |
0.40 |
|
|
|
|
rs935053 |
|
|
|
|
|
| AA |
172 (22.3) |
293 (25.4) |
Reference |
|
|
| AG |
373 (48.3) |
601 (52) |
1.10 |
0.87–1.40 |
|
| GG |
228 (29.5) |
259 (22.5) |
1.48 |
1.13–1.94 |
0.009 |
| AG+GG |
601 (78 ) |
860 (74.6) |
1.22 |
0.97–1.53 |
0.08 |
| A allele |
717 (46.4) |
1187 (51.5) |
Reference |
|
|
| G allele |
829 (53.6) |
1119 (48.5) |
1.22 |
1.06–1.39 |
0.004 |
| MAF |
0.54 |
0.49 |
|
|
|
|
rs10757257 |
|
|
|
|
|
| GG |
320 (41.3) |
407 (35.4) |
Reference |
|
|
| GA |
360 (46.5) |
567 (49.3) |
0.83 |
0.68–1.02 |
|
| AA |
94 (12.1) |
177 (15.4) |
0.65 |
0.48–0.88 |
0.02 |
| GA+AA |
454 (58.7) |
744 (64.6) |
0.79 |
0.65–0.96 |
0.02 |
| G allele |
1000 (64.6) |
1381 (60) |
Reference |
|
|
| A allele |
548 (35.4) |
921 (40) |
0.81 |
0.71–0.94 |
0.004 |
| MAF |
0.35 |
0.40 |
|
|
|
|
rs7023329 |
|
|
|
|
|
| AA |
230 (29.7) |
290 (25.2) |
Reference |
|
|
| AG |
377 (48.7) |
576 (50) |
0.84 |
0.67–1.06 |
|
| GG |
167 (21.6) |
287 (24.9) |
0.74 |
0.56–0.96 |
0.08 |
| AG+GG |
544 (70.3) |
863 (74.9) |
0.81 |
0.65–1.00 |
0.05 |
| A allele |
837 (54.1) |
1156 (50.1) |
Reference |
|
|
| G allele |
711 (45.9) |
1150 (49.9) |
0.86 |
0.75–0.98 |
0.02 |
| MAF |
0.46 |
0.50 |
|
|
|
|
rs10811629 |
|
|
|
|
|
| AA |
313 (40.4) |
395 (34.4) |
Reference |
|
|
| AG |
362 (46.8) |
579 (50.4) |
0.83 |
0.68–1.03 |
|
| GG |
99 (12.8) |
174 (15.2) |
0.70 |
0.52–0.95 |
0.05 |
| AG+GG |
461 (59.6) |
753 (65.6) |
0.80 |
0.66–0.98 |
0.03 |
| A allele |
988 (63.8) |
1369 (59.6) |
Reference |
|
|
| G allele |
560 (36.2) |
927 (40.4) |
0.84 |
0.73–0.97 |
0.02 |
| MAF |
0.36 |
0.40 |
|
|
|
|
rs1011970 |
|
|
|
|
|
| GG |
549 (71.3) |
850 (74.1) |
Reference |
|
|
| GT |
190 (24.7) |
277 (24.2) |
1.03 |
0.82–1.29 |
|
| TT |
31 (4) |
19 (1.7) |
2.24 |
1.22–4.13 |
0.04 |
| GT+TT |
221 (28.7) |
296 (25.8) |
1.11 |
0.89–1.38 |
0.35 |
| G allele |
1288 (83.6) |
1977 (86.3) |
Reference |
|
|
| T allele |
252 (16.4) |
315 (13.7) |
1.18 |
0.97- 1.42 |
0.10 |
| MAF |
0.17 |
0.14 |
|
|
|
|
CDKN2Atagging polymorphysms | |||||
|
SNP |
Cases (%) |
Controls (%) |
OR |
95% CI |
P value |
|
rs3731257 |
|
|
|
|
|
| GG |
398 (52) |
595 (51.8) |
Reference |
|
|
| GA |
302 (39.5) |
455 (39.6) |
0.94 |
0.77–1.15 |
|
| AA |
65 (8.5) |
98 (8.5) |
0.89 |
0.63–1.27 |
0.73 |
| GA+AA |
367 (48) |
553 (48.2) |
0.93 |
0.77–1.13 |
0.46 |
| G allele |
1098 (71.8) |
1645 (71.7) |
Reference |
|
|
| A allele |
432 (28.2) |
651 (28.4) |
0.94 |
0.81–1.09 |
0.43 |
| MAF |
0.28 |
0.28 |
|
|
|
|
rs11515 |
|
|
|
|
|
| GG |
538 (69.7) |
796 (69.6) |
Reference |
|
|
| GC |
214 (27.7) |
316 (27.7) |
0.96 |
0.78–1.19 |
|
| CC |
20 (2.6) |
31 (2.7) |
0.91 |
0.50–1.67 |
0.91 |
| GC +CC |
234 (30.3) |
347 (30.4) |
0.96 |
0.78–1.18 |
0.68 |
| G allele |
1290 (83.6) |
1908 (83.5) |
Reference |
|
|
| C allele |
254 (16.5) |
378 (16.5) |
0.96 |
0.80–1.15 |
0.66 |
| MAF |
0.17 |
0.17 |
|
|
|
|
rs3088440 |
|
|
|
|
|
| CC |
660 (85.4) |
1020 (90) |
Reference |
|
|
| CT |
108 (14) |
108 (9.5) |
1.52 |
1.13–2.05 |
|
| TT |
5 (0.7) |
5 (0.4) |
1.50 |
0.40–5.58 |
0.02 |
| CT+TT |
113 (14.6) |
113 (10) |
1.52 |
1.14–2.04 |
0.005 |
| C allele |
1428 (92.4) |
2148 (94.8) |
Reference |
|
|
| T allele |
118 (7.6) |
118 (5.2) |
1.48 |
1.12–1.96 |
0.005 |
| MAF |
0.08 |
0.05 |
|
|
|
|
rs2518719 |
|
|
|
|
|
| AA |
541 (70.8) |
808 (70.4) |
Reference |
|
|
| AG |
206 (27) |
320 (27.9) |
1.04 |
0.84–1.30 |
|
| GG |
17 (2.2) |
19 (1.7) |
1.58 |
0.79–3.19 |
0.42 |
| AG+GG |
223 (29.2) |
339 (29.6) |
1.07 |
0.87–1.33 |
0.51 |
| A allele |
1288(84.3) |
1936 (84.4) |
Reference |
|
|
| G allele |
240 (15.7) |
358 (15.6) |
1.09 |
0.91–1.31 |
0.36 |
| MAF |
0.16 |
0.16 |
|
|
|
|
rs2811708 |
|
|
|
|
|
| GG |
369 (46.8) |
563 (49.1) |
Reference |
|
|
| GT |
332 (43) |
475 (41.5) |
1.09 |
0.89–1.33 |
|
| TT |
71 (9.2) |
108 (9.4) |
1.05 |
0.74–1.48 |
0.70 |
| GT + TT |
403 (52.2) |
583 (50.9) |
1.08 |
0.89–1.31 |
0.42 |
| G allele |
1070 (69.3) |
1601 (69.9) |
Reference |
|
|
| T allele |
474 (30.7) |
691 (30.2) |
1.05 |
0.91–1.22 |
0.52 |
| MAF |
0.31 |
0.30 |
|
|
|
|
rs3731239 |
|
|
|
|
|
| AA |
356 (46.8) |
494 (43) |
Reference |
|
|
| AG |
318 (41.8) |
536 (46.6) |
0.84 |
0.68–1.03 |
|
| GG |
86 (11.3) |
120 (10.4) |
1.08 |
0.78–1.50 |
0.13 |
| AG+GG |
404 (53.2) |
656 (57) |
0.88 |
0.73–1.07 |
0.20 |
| A allele |
1030 (67.8) |
1524 (66.3) |
Reference |
|
|
| G allele |
490 (32.2) |
776 (33.7) |
0.97 |
0.84–1.12 |
0.65 |
| MAF |
0.33 |
0.34 |
|
|
|
|
rs2811710 |
|
|
|
|
|
| CC |
286 (37) |
450 (39.3) |
Reference |
|
|
| CT |
355 (45.9) |
542 (47.3) |
1.03 |
0.82–1.27 |
|
| TT |
132 (17.1) |
154 (13.4) |
1.40 |
1.05–1.87 |
0.06 |
| CT+TT |
487 (63) |
696 (60.7) |
1.10 |
0.91–1.35 |
0.30 |
| C allele |
927 (60) |
1442 (62.9) |
Reference |
|
|
| T allele |
619 (40) |
850 (37.1) |
1.15 |
1.00–1.32 |
0.05 |
| MAF |
0.40 |
0.37 |
|
|
|
|
rs3218020 |
|
|
|
|
|
| GG |
254 (32.8) |
381 (33.2) |
Reference |
|
|
| GA |
375 (48.5) |
560 (48.8) |
0.97 |
0.78–1.20 |
|
| AA |
145 (18.7) |
206 (17.9) |
0.96 |
0.73–1.27 |
0.95 |
| GA+AA |
520 (67.2) |
766 (66.8) |
0.97 |
0.79–1.18 |
0.74 |
| G allele |
883 (57) |
1322 (57.6) |
Reference |
|
|
| A allele |
665 (43) |
972 (42.4) |
0.98 |
0.85–1.12 |
0.76 |
| MAF |
0.43 |
0.42 |
|
|
|
|
rs3218009 |
|
|
|
|
|
| CC |
650 (84) |
965 (83.8) |
Reference |
|
|
| CG |
118 (15.3) |
180 (15.6) |
1.01 |
0.77–1.31 |
|
| GG |
6 (0.8) |
6 (0.5) |
1.96 |
0.61–6.29 |
0.52 |
| CG+GG |
124 (16) |
186 (16.2) |
1.04 |
0.80–1.34 |
0.79 |
| C allele |
1418 (91.6) |
2110 (91.7) |
Reference |
|
|
| G allele |
130 (8.4) |
192 (8.3) |
1.06 |
0.83–1.35 |
0.63 |
| MAF |
0.08 |
0.08 |
|
|
|
|
rs3217992 |
|
|
|
|
|
| CC |
229 (29.9) |
341 (29.7) |
Reference |
|
|
| CT |
380 (49.7) |
565 (49.2) |
0.95 |
0.76–1.19 |
|
| TT |
156 (20.4) |
243 (21.2) |
0.89 |
0.68–1.17 |
0.72 |
| CT+TT |
536 (70.1) |
808 (70.3) |
0.94 |
0.76–1.15 |
0.53 |
| C allele |
838 (54.8) |
1247 (54.3) |
Reference |
|
|
| T allele |
692 (45.2) |
1051 (45.7) |
0.95 |
0.83–1.08 |
0.42 |
| MAF |
0.45 |
0.46 |
|
|
|
|
rs1063192 |
|
|
|
|
|
| AA |
316 (40.9) |
445 (38.8) |
Reference |
|
|
| AG |
365 (47.3) |
537 (46.9) |
0.99 |
0.80–1.22 |
|
| GG |
91 (11.8) |
164 (14.3) |
0.89 |
0.65–1.20 |
0.72 |
| AG+GG |
456 (59.1) |
701 (61.2) |
0.97 |
0.79–1.17 |
0.72 |
| A allele |
997 (64.6) |
1427 (62.3) |
Reference |
|
|
| G allele |
547 (35.4) |
865 (37.7) |
0.95 |
0.83–1.10 |
0.51 |
| MAF |
0.35 |
0.38 |
|
|
|
|
rs573687 |
|
|
|
|
|
| GG |
380 (49.3) |
532 (46.4) |
Reference |
|
|
| GA |
329 (42.7) |
501 (43.7) |
0.95 |
0.78–1.17 |
|
| AA |
62 (8) |
114 (9.9) |
0.86 |
0.61–1.22 |
0.68 |
| GA+AA |
391 (50.7) |
615 (53.6) |
0.94 |
0.77–1.14 |
0.50 |
| G allele |
1089 (70.6) |
1565 (68.2) |
Reference |
|
|
| A allele |
453 (29.4) |
729 (31.8) |
0.94 |
0.81–1.09 |
0.40 |
| MAF |
0.29 |
0.32 |
|
|
|
|
rs13298881 |
|
|
|
|
|
| TT |
565 (73.8) |
862 (75.2) |
Reference |
|
|
| TC |
183 (23.9) |
262 (22.8) |
1.14 |
0.91–1.43 |
|
| CC |
18 (2.4) |
23 (2) |
1.26 |
0.65–2.45 |
0.46 |
| TC+CC |
201 (26.2) |
285 (24.9) |
1.15 |
0.92–1.43 |
0.22 |
| T allele |
1313 (85.7) |
1986 (86.6) |
Reference |
|
|
| C allele |
219 (14.3) |
308 (13.4) |
1.14 |
0.93–1.38 |
0.21 |
| MAF |
0.14 |
0.13 |
|
|
|
|
rs545226 |
|
|
|
|
|
| AA |
207 (26.8) |
300 (26.1) |
Reference |
|
|
| AG |
378 (48.9) |
577 (50.3) |
0.88 |
0.70–1.11 |
|
| GG |
188 (24.3) |
271 (23.6) |
0.93 |
0.71–1.22 |
0.55 |
| AG+GG |
566 (73.2) |
848 (73.9) |
0.90 |
0.72–1.11 |
0.32 |
| A allele |
792 (51.2) |
1177 (51.3) |
Reference |
|
|
| G allele |
754 (48.8) |
1119 (48.7) |
0.96 |
0.84–1.10 |
0.57 |
| MAF |
0.49 |
0.49 |
|
|
|
|
rs10811640 |
|
|
|
|
|
| GG |
184 (24.1) |
298 (26.1) |
Reference |
|
|
| GT |
371 (48.5) |
565 (49.4) |
1.02 |
0.81–1.29 |
|
| TT |
210 (27.5) |
280 (24.5) |
1.12 |
0.86–1.46 |
0.68 |
| GT+TT |
581 (76) |
845 (73.9) |
1.05 |
0.84–1.32 |
0.65 |
| G allele |
739 (48.3) |
1161 (50.8) |
Reference |
|
|
| T allele |
791 (51.7) |
1125 (49.2) |
1.06 |
0.92–1.21 |
0.42 |
| MAF | 0.52 | 0.49 | |||
Values in bold indicate statistical significance.
The polymorphisms rs4636294, rs2218220 and rs935053 linked with an r2 =1 on HapMap data; showed a similar pattern of association with melanoma risk. All the three variants are located in the intergenic region between IFNE and MTAP genes (Figure 1). The variant A-allele and the homozygous carriers of the variant allele of the rs4636294 polymorphism showed an increased risk of melanoma with an OR of 1.19 (95% CI 1.04-1.36) and OR of 1.41 (95% CI 1.08-1.85), respectively. Same pattern of association was observed for the rs2218220 and rs935053 polymorphisms (Table 1). According to HapMap data 117 SNPs (including the rs2218220 and rs935053) were tagged by the rs4636294 and all were located in intergenic region between IFNE and MTAP genes.
Linkage disequilibrium analysis using HapMap data showed that the rs1335510 and rs1341866 polymorphisms also located in the intergenic region between IFNE and MTAP genes were in complete linkage disequilibrium with an r2 =1; an additional polymorphism rs10757257 located in intron 1 of MTAP gene was in linkage disequilibrium with r2 =0.89 with rs1335510 and rs1341866 (Figure 1). The polymorphism rs1335510 showed a statistically significant decreased risk for the carriers of the G-allele (OR 0.76, 95% CI 0.63-0.93). A similar trend was observed for the carriers of the variant C-allele of the polymorphism rs1341866 and the carriers of the variant A-allele of rs10757257 (Table 1). Among the 37 variants tagged (with r2 ≥0.8) by rs1335510 (including rs1341866 and rs10757257), one was located in exon 3 of MTAP gene and the other variants were located in intergenic regions or were intronic to MTAP. The regions defined by rs751173 and rs4636294-rs2218220-rs935053 were linked to each other with an r2 of 0.7. The regions defined by rs751173 and rs1335510-rs1341866 were linked with an r2 of 0.41. And finally the regions defined by rs4636294-rs2218220-rs935053 and rs1335510-rs1341866 were in linkage disequilibrium with an r2 of 0.65.
The polymorphism rs7023329 located in intron 2 of MTAP that tagged 10 additional variants showed a statistical significant association with decreased risk of melanoma (Figure 1). The carriers of the variant G-allele were at a decreased risk of melanoma with an OR 0.81 (95% CI 0.65-1.00; Table 1). A statistically significant decreased risk of melanoma was also observed for the carriers of the G-variant of the polymorphism rs10811629 located in intron 5 of MTAP gene (OR 0.80, 95% CI 0.66-0.98) (Table 1).
No statistically significant association with melanoma was observed for the carrier of the variant T-allele of the rs1011970 polymorphism located in intron 9 of ANRIL gene (Figure 1). However, a statistically significant association with increased melanoma risk was observed for the homozygous carriers of the variant T-allele of the rs1011970 polymorphism (OR 2.24, 95% CI 1.22-4.13; Table 1).
The polymorphism rs3088440 located at the 3’UTR of CDKN2A gene, tagged six variants located in intronic regions of CDKN2A/ARF, ANRIL and 3’UTR of CDKNB gene as well as intergenic variants between MTAP and CDKN2A genes. The carriers of the variant T-allele of the rs3088440 variant showed a statistically significant increased risk of melanoma with an OR of 1.52 (95% CI 1.14-2.04) (Figure 2; Table 1). In addition statistically significant association with melanoma risk was also observed for the variant T-allele of the polymorphism rs2811710 located in intron 1 of CDKN2A/ARF (OR 1.15, 95% CI 1.00-1.32) (Figure 2; Table 1). The use of logistic regression model that, besides all covariates like age and gender, included all the 25 genotyped polymorphisms on chromosome 9p21 showed retention of statistical significant association for the rs935053 and rs3088440 polymorphisms.
Associated variants and expression quantitative trait loci analysis on chromosome 9p21
All the associated polymorphisms and tagged SNPs were analyzed for being eQTL. Polymorphisms that tagged to rs751173, rs4636294 (and the linked rs2218220 and rs935053), rs1335510 (and the linked rs1341866 and rs10757257), rs7023329 and rs2811710 were reported as eQTL. All the eQTL variants were cis acting. The genes with affected expressions were MTAP and CDKN2B (Additional file 3: Table S3).
Linkage disequilibrium analysis
Linkage disequilibrium analysis was performed in the Spanish population (cases and controls were analyzed together). We observed that the linkage disequilibrium with r2 ≥0.8 reported in HapMap was replicated in our data for rs4636294, rs2218220 and rs935053 as well as for rs1335510, rs1341866 and rs10757257 variants. An additional linkage disequilibrium not reported in HapMap data was observed between the rs3218020 and rs3217992. The linkage disequilibrium between the two variants was r2 of 0.86 (Figure 3).
Figure 3.
Linkage disequilibrium map of the genotyped polymorphisms on chromosome 9p21 in the Spanish population. *CDKN2A isoform 1 encodes p16INK4A ;**CDKN2A isoform 4 (CDKN2A/ARF) encodes p14ARF.
Interaction of associated variants with MC1R variants and host risk factors
Gene-gene interaction analysis was performed only for the polymorphisms that showed statistically significant association with melanoma risk. Interaction analysis with MC1R variants did not showed any statistically significant association (Additional file 4: Table S4). A statistical significant interaction for melanoma risk was observed between phototypes and the rs10811629 (Additional file 5: Table S5). Strongest interaction was observed between the homozygotes for the A–allele and phototype II with OR of 15.93 (95% CI 5.34-47.54) when compared to the carrier of the variant G-allele with phototype V. A statistically significant interaction was also observed between carrier of the A-allele and phototype I (OR 8.43; 95% CI 2.51-28.33); phototype III (OR 5.83; 95% CI 1.99-17.11) and phototype IV (OR 7.79; 95% CI 2.59-23.43; Additional file 5: Table S5). No other statistically significant interaction was observed between the genotyped polymorphisms and other host risk factors.
The strongest gene-host factor interaction observed between homozygotes for the A–allele and phototype II was also tested for departure from additivity and multiplicativity. No departure from additivity (ICR 9.07; 95% CI −1.54-32.81; P value 0.09) and multiplicativity was observed (MII 1.97; 95% CI 0. 67–5.85; P value 0.15).
Discussion
Several GWAS over the years have shown association between the 9p21 locus and several cancers including glioma, basal cell carcinoma of skin and several diseases like type-2 diabetes (Figure 4) [20-31]. In particular two GWAS reported the association of nine variants with one additional variant in the replication phase with risk of either cutaneous nevi or melanoma or both [8,9]. In the present study we could replicate the association between the variants reported in GWAS with melanoma risk. In addition we also genotyped 15 variants tagging the CDKN2A gene and we observed a statistically significant association with melanoma risk for the previously inconsistently associated rs3088440 variant.
Figure 4.
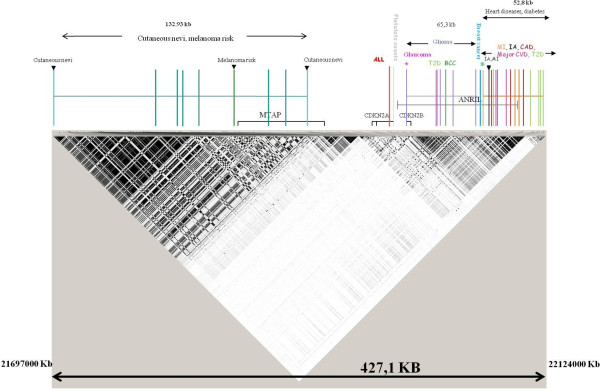
Linkage disequilibrium map of chromosome 9p21 from 21697000 bp to 22124000 bp. The map derived from HapMap (release #27) shows the SNPs associated in GWAS with several cancers including melanoma and other diseases. T2D Type II diabetes, MI myocardialinfarction, CVD cardiovascular disease, CAD coronary artery disease, BCC Basal cell carcinoma, IA Intracranial aneurism, ALL Acute lymphoblastic leukemia.
All the variants selected from the two GWAS associated with melanoma risk excluding rs1011970 were located in a 132.9 Kb region including MTAP gene. In this region the associated variants and the 187 tagged variants were located in intergenic region telomeric to MTAP gene or intronic to MTAP gene; only one variant was located in exon 3 of MTAP. Interestingly a study reported that the MTAP variant rs10757257 was associated with increased risk of superficial spreading/nodular melanoma but not with lentigo malignant melanoma that is the subtype associated with chronic sun exposure. Those results suggested that the relationship between MTAP and melanoma could be subtype-specific [32]. MTAP gene plays an important role in polyamine metabolism and the salvage of adenine and methionine. MTAP is reported to show low expression in malignant melanoma cells and conversely its substrate i.e., 5′-deoxy-5’-(methylthio) adenosine (MTA) has higher expression in melanoma cells compared to normal melanocytes. The increased level of MTA has been related to an increased invasive potential and vascuologenesis [33,34]. Loss of MTAP expression in melanoma is mainly due to hypermethylation of the promoter region. It was suggested that loss of MTAP expression in malignant melanomas might have an impact on therapeutic success by compromising tumor response to interferon treatment through its impact on STAT1 activity [33,35,36].
On the basis of the allelic P values we observed that all the GWAS variants except rs1011970 (intron 9 ANRIL) showed statistically significant association. The polymorphism rs1011970 however showed statistically significant association with melanoma risk only for the homozygotes of the variant T-allele. The polymorphism rs1011970 reported to be associated with melanoma was also associated with breast cancer risk modulation in a GWAS [9,37]. ANRIL gene located in the antisense orientation to that of the CDKN2A and CDKN2B genes consists of 19 exons. ANRIL gene is transcribed into several different splice variants of non-coding-RNA [38-40]. The first exon of ANRIL gene overlaps the two exons of CDKN2B gene. A study observed that silencing of ANRIL resulted in 8-fold increase of CDKN2B expression suggesting a negative regulation. The study suggested that ANRIL binds to and recruits polycomb repression complex 2 (PCR2) to repression of CDKN2B[41]. In addition, the 5’ end of the first exon of the ANRIL gene is located ~300 bp upstream of the transcription start site of CDKN2A/ARF gene [39]. The two genes are physically linked by the intergenic region and co-regulated by E2F1 [40]. Interestingly several GWAS identified the association of genetic variants in the ANRIL gene with several cancer and diseases including basal cell carcinoma, breast cancer, glioma, coronary disease, intracranial aneurysm and also type 2 diabetes [39]. The association of variants within ANRIL gene and several cancers suggests a plausible involvement of the gene in melanoma etiology. The role of CDKN2A on melanogenesis is mainly explained by the presence of germline mutations in a proportion of familial melanoma [7]. However, the influence of the polymorphisms rs11515 (500 G>C), rs3088440 (540 C>T) at the 3′ UTR of the gene and the coding variant A148T located in exon 2 on melanoma risk has been extensively investigated in different studies over the years by candidate gene approach and meta-analysis. The association of the CDKN2A polymorphisms with melanoma risk reported in those studies lack consistency [14-17,42,43]. In the present study we observed for the carriers of the T –allele of the polymorphism rs3088440 a statistically significant association with melanoma risk. The associated variant tagged a total of 6 SNPs of which 3 were located in intergenic region and the other three in intron 1 of CDKN2A, 3’UTR of CDKN2B and intron 3 of ANRIL. Association with melanoma risk was also observed for the variant T-allele of rs2811710 located in intron 1 of CDKN2A/ARF. The associated variant tagged (with r2 ≥0.80) three polymorphisms, all located in a intergenic region between the MTAP and CDKN2A genes. The effect of the rs3088440 at the 3′UTR and of the intronic variants on CDKN2A cannot be ruled out.
The majority of the variants associated with melanoma risk and also most of the tagged variants in the present study were located in intergenic or intronic regions. The role of the associated and tagged variants in the melanoma etiology still needs to be clarified. However the intronic variants can possibly affect the expression of respective genes as being part of enhancer elements through binding of transcription factors as reported in previous studies [44,45]. Intronic variants can also affect gene splicing. Although some intronic polymorphisms are not located at the splice junctions, they can still act so as to change the splicing phenotype as a consequence of their being located within an intron splice enhancer or branchpoint site, or by activating a cryptic splice site as reported in several studies [46]. Alteration in gene splicing and consequent alteration in protein structure could lead to disease development [47]. Intronic and intergenic polymorphisms can be expression quantitative trait loci (eQTLs) and can alter gene expression possibly explaining the observed association with melanoma risk. Moreover, the possibility of the associated variants being in linkage disequilibrium with rare functional variants cannot be precluded.
We investigated the associated and linked variants to be eQTL using the eQTL browser eqtl.uchicago.edu/Home.html. We observed that variants linked to the associated intergenic rs751173, rs4636294, rs2218220, rs935053, rs1335510 and the intronic rs7023329 and rs2811710 polymorphisms were eQTL and showed regulation of MTAP and CDKN2B genes. CDKN2B gene encodes for the tumor suppressor protein p15INK4B that it is homologous to p16INK4A[48]. Several lines of evidence indicate that p15INK4B inactivation is not significant in the development of melanoma. First, despite its frequent co-deletion with CDKN2A, germline or acquired mutations that target CDKN2B exclusively have not been observed [49,50]. Second, CDKN2B-null mice showed a minimal cancer-prone phenotype and do not develop melanoma [49,51].
In the present study we observed effect of the associated variants on chromosome 9p21 independent of MC1R variants on melanoma risk. However due to the limited size of the present study we could not exclude a possible epistatic interaction of MC1R variants and 9p21 locus variants on melanoma risk. We also investigated presence of epistatic interaction between associated variants and host risk factor and we observed an interaction between phototype and the rs10811629 (intron 5 MTAP) variant. The highest increased risk was observed for the homozygotes of the risk A-allele with phototype II compared to the reference carrier of the G-allele and phototype V. No other epistatic interaction was observed in the gene-host risk factors analysis. It may be pointed out that self-reporting of phenotype in controls could be of questionable precision, therefore the interpretation of the detected association between the variant and the phototype desires a cautious interpretation.
Conclusions
In conclusion, the present study on variants at 9p21 locus replicated the association with melanoma risk of the GWAS selected variants. In addition for the variants selected for tagging CDKN2A gene; the strongest association with melanoma risk was observed for rs3088440 (3′UTR of CDKN2A gene) that has been inconsistently associated with melanoma in previous studies [14-16]. In the present study we also observed an epistatic interaction between the polymorphism rs10811629 (intron 5 MTAP) and phototype. The effect of the associated or tagged variants on the respective genes and their role as eQTL on other genes could not be ruled out. However it is biologically plausible that variants affecting the genes MTAP, CDKN2A and ANRIL could have an important role in melanoma etiology. Further investigations are needed to clarify the role of CDKN2B in melanoma disease and how the variants affecting the gene could be related to melanoma risk. Our finding together with previous GWAS provided evidence of an involvement of variants at the 9p21 locus and melanoma risk. This represents a unique study that investigated a large homogenous case–control population for the association of tagging polymorphisms on chromosome 9p21 discovered through genome wide association studies and polymorphisms at the CDKN2A locus with melanoma risk and association with host factors.
Abbreviations
CDKN2A: Cyclin-dependent kinase inhibitor 2A; GWAS: Genome wide association studies; CDK4: Cyclin-dependent kinase 4; Rb: Retinoblastoma; MDM2: Mouse double minute −2; MTAP: Methylthioadenosine phosphorylase; SNP: Single nucleotide polymorphism; ANRIL: Antisense non coding RNA; CDKN2B: Cyclin-dependent kinase inhibitor 2B; AJCC: American Joint Committee on Cancer; OR: Odds ratios; 95% CI: 95% confidence intervals; LTR: Likelihood ratio tests; MC1R: Melanocortin receptor 1; eQTL: expression quantitative trait loci; IFNE: Interferon, epsilon; PCR2: Polycomb repression complex 2; E2F1: E2F transcription factor 1; MTA: 5’-deoxy-5’-(methylthio) adenosine; STAT1: Signal transducer and activator of transcription 1.
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
LM genotyped and performed statistical analysis of the polymorphisms and wrote the paper. PSR extracted the DNA from blood for the healthy donors and genotyped. JLB supervised statistical analysis for gene-phenotypic traits interaction. DP and CR provided samples and data. KH supervised the study. EN provided the samples for the study and clinical data. RK supervised and coordinated the study; revised the manuscript. All authors contributed to the manuscript finalization. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Detailed information of genotyped single nucleotide polymorphisms on chromosome 9p21.
Primer sequences and conditions for PCR.
eQTL for the polymorphisms tagged (r2 ≥0.8) by rs751173, rs4636294, rs2218220, rs935053, rs1335510, rs7023329, rs2811710.
Effect of interaction between the polymorphisms rs751173, rs4636294, rs2218220, rs1335510, rs1341866, rs935053, rs10757257, rs7023329, rs10811629, rs1011970, rs3088440, rs2811710 and MC1R variants on melanoma risk.
Effect of interaction between the polymorphisms rs751173, rs4636294, rs2218220, rs1335510, rs1341866, rs935053, rs10757257, rs7023329, rs10811629, rs1011970, rs3088440, rs2811710 and phenotypic traits on melanoma risk.
Contributor Information
Livia Maccioni, Email: livia.mac@libero.it.
Panduranga Sivaramakrishna Rachakonda, Email: s.rachakonda@dkfz.de.
Justo Lorenzo Bermejo, Email: lorenzo@imbi.uni-heidelberg.de.
Dolores Planelles, Email: planelles_dol@gva.es.
Celia Requena, Email: celiareq@hotmail.com.
Kari Hemminki, Email: K.Hemminki@dkfz-heidelberg.de.
Eduardo Nagore, Email: eduyame@meditex.es.
Rajiv Kumar, Email: r.kumar@dkfz.de.
References
- Garbe C, Blum A. Epidemiology of cutaneous melanoma in Germany and worldwide. Skin Pharmacol Appl Skin Physiol. 2001;14(5):280–290. doi: 10.1159/000056358. [DOI] [PubMed] [Google Scholar]
- Berwick M, Wiggins C. The current epidemiology of cutaneous malignant melanoma. Front Biosci. 2006;11:1244–1254. doi: 10.2741/1877. [DOI] [PubMed] [Google Scholar]
- Maddodi N, Setaluri V. Role of UV in cutaneous melanoma. Photochem Photobiol. 2008;vol. 84(2008/02/06 edn):528–536. doi: 10.1111/j.1751-1097.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- Gallagher RP, Spinelli JJ, Lee TK. Tanning beds, sunlamps, and risk of cutaneous malignant melanoma. Cancer Epidemiol Biomarkers Prev. 2005;14(3):562–566. doi: 10.1158/1055-9965.EPI-04-0564. [DOI] [PubMed] [Google Scholar]
- MacKie RM, Hauschild A, Eggermont AM. Epidemiology of invasive cutaneous melanoma. Ann Oncol. 2009;20(Suppl 6):vi1–vi7. doi: 10.1093/annonc/mdp252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Colditz GA, Hunter DJ. Risk factors for skin cancers: a nested case–control study within the Nurses’ health study. Int J Epidemiol. 2006;35(6):1514–1521. doi: 10.1093/ije/dyl197. [DOI] [PubMed] [Google Scholar]
- Hansson J. Familial cutaneous melanoma. Adv Exp Med Biol. 2010;685:134–145. doi: 10.1007/978-1-4419-6448-9_13. [DOI] [PubMed] [Google Scholar]
- Falchi M, Bataille V, Hayward NK, Duffy DL, Bishop JA, Pastinen T, Cervino A, Zhao ZZ, Deloukas P, Soranzo N. et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat Genet. 2009;41(8):915–919. doi: 10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DT, Demenais F, Iles MM, Harland M, Taylor JC, Corda E, Randerson-Moor J, Aitken JF, Avril MF, Azizi E. et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41(8):920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI, Wang LE, Lee JE, Gershenwald JE, Chen WV, Fang S, Kosoy R, Zhang M, Qureshi AA, Vattathil S. et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum Mol Genet. 2011;20(24):5012–5023. doi: 10.1093/hmg/ddr415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor S, Montgomery GW, Liu JZ, Zhao ZZ, Henders AK, Stark M, Schmid H, Holland EA, Duffy DL, Zhang M. et al. Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3. Nat Genet. 2011;43(11):1114–1118. doi: 10.1038/ng.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JH, Iles MM, Harland M, Taylor JC, Aitken JF, Andresen PA, Akslen LA, Armstrong BK, Avril MF, Azizi E. et al. Genome-wide association study identifies three new melanoma susceptibility loci. Nat Genet. 2011;43(11):1108–1113. doi: 10.1038/ng.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken J, Welch J, Duffy D, Milligan A, Green A, Martin N, Hayward N. CDKN2A variants in a population-based sample of Queensland families with melanoma. J Natl Cancer Inst. 1999;91(5):446–452. doi: 10.1093/jnci/91.5.446. [DOI] [PubMed] [Google Scholar]
- Kumar R, Smeds J, Berggren P, Straume O, Rozell BL, Akslen LA, Hemminki K. A single nucleotide polymorphism in the 3'untranslated region of the CDKN2A gene is common in sporadic primary melanomas but mutations in the CDKN2B, CDKN2C, CDK4 and p53 genes are rare. Int J Cancer. 2001;95(6):388–393. doi: 10.1002/1097-0215(20011120)95:6<388::AID-IJC1069>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Debniak T, Scott RJ, Huzarski T, Byrski T, Rozmiarek A, Debniak B, Zaluga E, Maleszka R, Kladny J, Gorski B. et al. CDKN2A common variants and their association with melanoma risk: a population-based study. Cancer Res. 2005;65(3):835–839. [PubMed] [Google Scholar]
- Ibarrola-Villava M, Fernandez LP, Pita G, Bravo J, Floristan U, Sendagorta E, Feito M, Aviles JA, Martin-Gonzalez M, Lazaro P. et al. Genetic analysis of three important genes in pigmentation and melanoma susceptibility: CDKN2A, MC1R and HERC2/OCA2. Exp Dermatol. 2010;19(9):836–844. doi: 10.1111/j.1600-0625.2010.01115.x. [DOI] [PubMed] [Google Scholar]
- Chatzinasiou F, Lill CM, Kypreou K, Stefanaki I, Nicolaou V, Spyrou G, Evangelou E, Roehr JT, Kodela E, Katsambas A. et al. Comprehensive field synopsis and systematic meta-analyses of genetic association studies in cutaneous melanoma. J Natl Cancer Inst. 2011;103(16):1227–1235. doi: 10.1093/jnci/djr219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer D, Nagore E, Bermejo JL, Figl A, Botella-Estrada R, Thirumaran RK, Angelini S, Hemminki K, Schadendorf D, Kumar R. Melanocortin receptor 1 variants and melanoma risk: a study of 2 European populations. Int J Cancer. 2009;125(8):1868–1875. doi: 10.1002/ijc.24548. [DOI] [PubMed] [Google Scholar]
- Maccioni L, Rachakonda PS, Scherer D, Bermejo JL, Planelles D, Requena C, Hemminki K, Nagore E, Kumar R. Variants at chromosome 20 (ASIP locus) and melanoma risk. Int J Cancer. 2013;132(1):42–54. doi: 10.1002/ijc.27648. [DOI] [PubMed] [Google Scholar]
- Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY. et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41(8):899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S. et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, Voight BF, Lyssenko V, Burtt NP, De Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ. et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU. et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpson NJ, Lindgren CM, Weedon MN, Randall J, Ouwehand WH, Strachan DP, Rayner NW, Walker M, Hitman GA, Doney AS. et al. Adiposity-related heterogeneity in patterns of type 2 diabetes susceptibility observed in genome-wide association data. Diabetes. 2009;58(2):505–510. doi: 10.2337/db08-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, De Bakker PI, Abecasis GR, Almgren P, Andersen G. et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40(5):638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F, Serizawa M, Yamamoto K, Fujisawa T, Nakashima E, Ohnaka K, Ikegami H, Sugiyama T, Katsuya T, Miyagishi M. et al. Confirmation of multiple risk Loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes. 2009;58(7):1690–1699. doi: 10.2337/db08-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G. et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42(7):579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G. et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- Sherborne AL, Hosking FJ, Prasad RB, Kumar R, Koehler R, Vijayakrishnan J, Papaemmanuil E, Bartram CR, Stanulla M, Schrappe M. et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet. 2010;42(6):492–494. doi: 10.1038/ng.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey SN, Sulem P, Masson G, Gudjonsson SA, Thorleifsson G, Jakobsdottir M, Sigurdsson A, Gudbjartsson DF, Sigurgeirsson B, Benediktsdottir KR. et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41(8):909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MG, Atwood LD, Benjamin EJ, Cupples LA, D’Agostino RB Sr, Fox CS, Govindaraju DR, Guo CY, Heard-Costa NL, Hwang SJ. et al. Framingham Heart Study 100K project: genome-wide associations for cardiovascular disease outcomes. BMC Med Genet. 2007;8(Suppl 1):S5. doi: 10.1186/1471-2350-8-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvaskoff M, Whiteman DC, Zhao ZZ, Montgomery GW, Martin NG, Hayward NK, Duffy DL. Polymorphisms in nevus-associated genes MTAP, PLA2G6, and IRF4 and the risk of invasive cutaneous melanoma. Twin research and human genetics : the official journal of the International Society for Twin Studies. 2011;14(5):422–432. doi: 10.1375/twin.14.5.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrmann I, Wallner S, Komyod W, Heinrich PC, Schuierer M, Buettner R, Bosserhoff AK. Characterization of methylthioadenosin phosphorylase (MTAP) expression in malignant melanoma. Am J Pathol. 2003;163(2):683–690. doi: 10.1016/S0002-9440(10)63695-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens AP, Spangler B, Wallner S, Kreutz M, Dettmer K, Oefner PJ, Bosserhoff AK. Direct and tumor microenvironment mediated influences of 5'-deoxy-5'-(methylthio)adenosine on tumor progression of malignant melanoma. J Cell Biochem. 2009;106(2):210–219. doi: 10.1002/jcb.21984. [DOI] [PubMed] [Google Scholar]
- Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, David M. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104(5):731–741. doi: 10.1016/S0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- Wild PJ, Meyer S, Landthaler M, Hofstaedter F, Bosserhoff AK. A potential predictive marker for response to interferon in malignant melanoma. Journal der Deutschen Dermatologischen Gesellschaft = Journal of the German Society of Dermatology: JDDG. 2007;5(6):456–459. doi: 10.1111/j.1610-0387.2007.06303.x. [DOI] [PubMed] [Google Scholar]
- Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, Seal S, Ghoussaini M, Hines S, Healey CS. et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42(6):504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkersen L, Kyriakou T, Goel A, Peden J, Malarstig A, Paulsson-Berne G, Hamsten A, Hugh W, Franco-Cereceda A, Gabrielsen A. et al. Relationship between CAD risk genotype in the chromosome 9p21 locus and gene expression. Identification of eight new ANRIL splice variants. PLoS One. 2009;4(11):e7677. doi: 10.1371/journal.pone.0007677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25(2):444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- Sato K, Nakagawa H, Tajima A, Yoshida K, Inoue I. ANRIL is implicated in the regulation of nucleus and potential transcriptional target of E2F1. Oncol Rep. 2010;24(3):701–707. doi: 10.3892/or_00000910. [DOI] [PubMed] [Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spica T, Portela M, Gerard B, Formicone F, Descamps V, Crickx B, Ollivaud L, Archimbaud A, Dupin N, Wolkenstein P. et al. The A148T variant of the CDKN2A gene is not associated with melanoma risk in the French and Italian populations. J Invest Dermatol. 2006;126(7):1657–1660. doi: 10.1038/sj.jid.5700293. [DOI] [PubMed] [Google Scholar]
- Pjanova D, Engele L, Randerson-Moor JA, Harland M, Bishop DT, Newton Bishop JA, Taylor C, Debniak T, Lubinski J, Kleina R. et al. CDKN2A and CDK4 variants in Latvian melanoma patients: analysis of a clinic-based population. Melanoma Res. 2007;17(3):185–191. doi: 10.1097/CMR.0b013e328014a2cd. [DOI] [PubMed] [Google Scholar]
- Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Bjorklund M, Wei G, Yan J, Niittymaki I. et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41(8):885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- Visser M, Kayser M, Palstra RJ. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res. 2012;22(3):446–455. doi: 10.1101/gr.128652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN. Functional intronic polymorphisms: buried treasure awaiting discovery within our genes. Hum Genomics. 2010;4(5):284–288. doi: 10.1186/1479-7364-4-5-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AJ, Cooper TA. The pathobiology of splicing. J Pathol. 2010;220(2):152–163. doi: 10.1002/path.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloethner S, Scherer D, Drechsel M, Hemminki K, Kumar R. Malignant melanoma–a genetic overview. Actas Dermosifiliogr. 2009;100(Suppl 1):38–51. [PubMed] [Google Scholar]
- Chin L. The genetics of malignant melanoma: lessons from mouse and man. Nat Rev Cancer. 2003;3(8):559–570. doi: 10.1038/nrc1145. [DOI] [PubMed] [Google Scholar]
- Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998;1378(2):F115–177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- Latres E, Malumbres M, Sotillo R, Martin J, Ortega S, Martin-Caballero J, Flores JM, Cordon-Cardo C, Barbacid M. Limited overlapping roles of P15(INK4b) and P18(INK4c) cell cycle inhibitors in proliferation and tumorigenesis. EMBO J. 2000;19(13):3496–3506. doi: 10.1093/emboj/19.13.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed information of genotyped single nucleotide polymorphisms on chromosome 9p21.
Primer sequences and conditions for PCR.
eQTL for the polymorphisms tagged (r2 ≥0.8) by rs751173, rs4636294, rs2218220, rs935053, rs1335510, rs7023329, rs2811710.
Effect of interaction between the polymorphisms rs751173, rs4636294, rs2218220, rs1335510, rs1341866, rs935053, rs10757257, rs7023329, rs10811629, rs1011970, rs3088440, rs2811710 and MC1R variants on melanoma risk.
Effect of interaction between the polymorphisms rs751173, rs4636294, rs2218220, rs1335510, rs1341866, rs935053, rs10757257, rs7023329, rs10811629, rs1011970, rs3088440, rs2811710 and phenotypic traits on melanoma risk.