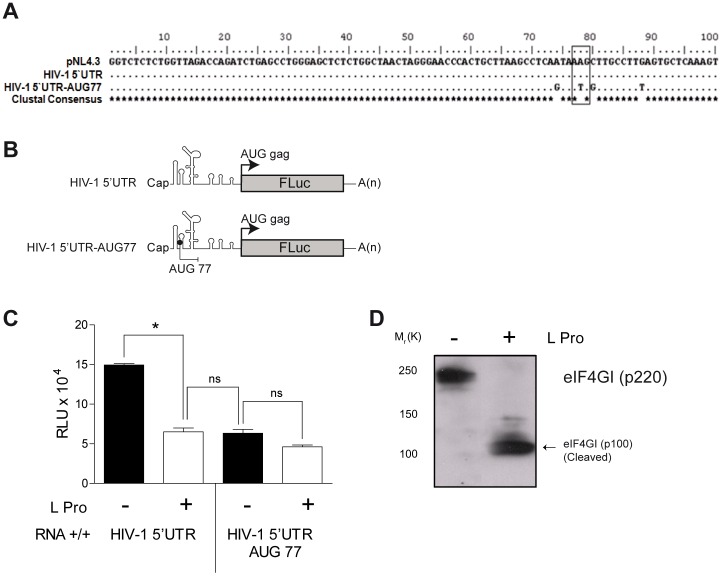
Figure 1. Contribution of the HIV-1 IRES to the overall translational activity displayed by the capped HIV-1 5′UTR.
(A) The partial sequence of the HIV-1 5′UTR from the proviral clone pNL4.3 (AF324493l) was aligned against the intercistronic region, HIV-1 5′UTR, recovered from the dl HIV-1 IRES plasmid [16] and the mutant HIV-1 5′UTR-AUG 77 in which an optimal initiation codon was inserted at position 77 with respect to the reference sequence. The block highlights the introduced initiation codon. (B) Schematic representation of the capped and polyadenylated monocistronic mRNAs used in this study. In the HIV-1 5′UTR monocistronic RNA the 5′UTR of the full-length HIV-1 mRNA was cloned upstream of the FLuc reporter. The reporter open reading frame (ORF) was left in frame with the Gag protein initiation codon (arrow). In the HIV-1 5′UTR AUG 77 monocistronic RNA and additional optimal initiation codon has been added at position 77 upstream of the putative internal ribosome entry site (IRES) and out of frame (end point line) with respect to the FLuc ORF. (C) In vitro transcribed capped and polyadenylated (+/+) RNA were translated in absence (−; black bar) or presence (+; white bar) of FMDV L protease [33]. Values are the means ± SEM from three independent experiments each conducted in triplicate. (D) Analysis of eIF4GI cleavage. RRL translation reactions (10 ul) without (lane 1) or with (lane 2) FMDV L protease (2% V/V) were resolved by SDS/gradient 5%–20% PAGE, transferred to nitrocellulose membrane, detected using polyclonal antibodies eIF4GI as previously described [33]. Statistical analysis was performed by Student t-test (*p<0.05). (B).