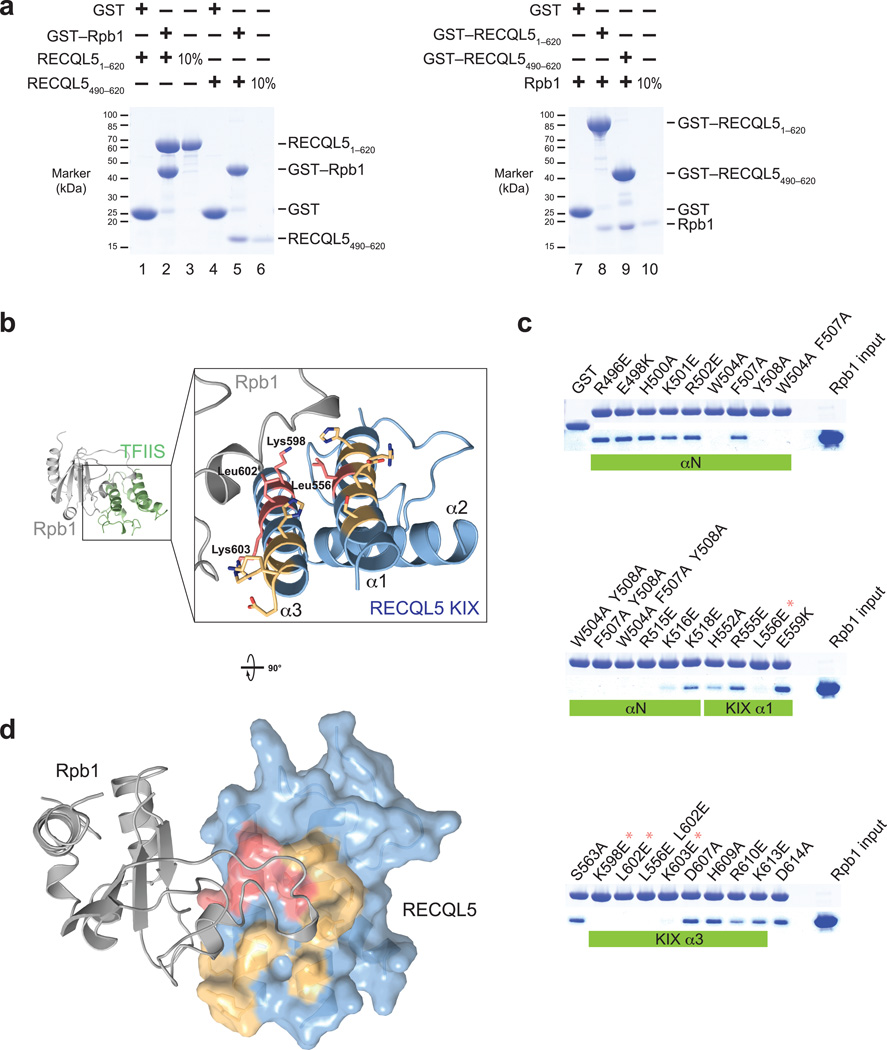
Figure 3. The RECQL5 IRI domain binds to a loop element in the Rpb1 jaw of Pol II.
(a) Pull–down assays of two different RECQL5 fragments and a recombinantly expressed Rpb1 fragment (residues 1168–1302). Left: Recombinant glutathione–S–transferase (GST)–tagged Rpb11168–1302 was immobilized on glutathione sepharose beads and incubated with RECQL51–620 or the RECQL5 IRI domain (residues 490–620). Bound proteins were analyzed by SDS–PAGE and visualized by Coomassie staining. Right: GST–fused RECQL51–620 or RECQL5490–620 were immobilized on glutathione sepharose beads and incubated with untagged recombinant Rpb11168–1302. (b) Model of the RECQL5 KIX–Rpb1 interaction surface based on the TFIIS–Pol II crystal structure, generated by superposition of the KIX domain and TFIIS domain II. Surface residues mutated in (c) are shown in stick representation. RECQL5 residues whose mutations abolish interaction with hRpb1 are highlighted in red, and mutated residues that have no effect on interaction with Rpb1 are shown in yellow. (c) Pull–down assays of Rpb11168–1302 by GST–RECQL5490–620 IRI domain mutants. Large hydrophobic residues important for interaction (W504, F507, Y508) are located in the first alpha–helix of the IRI domain (αN, not present in our crystal structure). Asterisks denote mutations that abolish interaction with Rpb1 and are part of the KIX domain (see b). (d) Model of the RECQL5 KIX–Rpb1 interaction. The KIX domain is depicted as a surface representation and colored as in (b). Mutations that abolish interaction cluster in a single patch on the KIX domain surface.