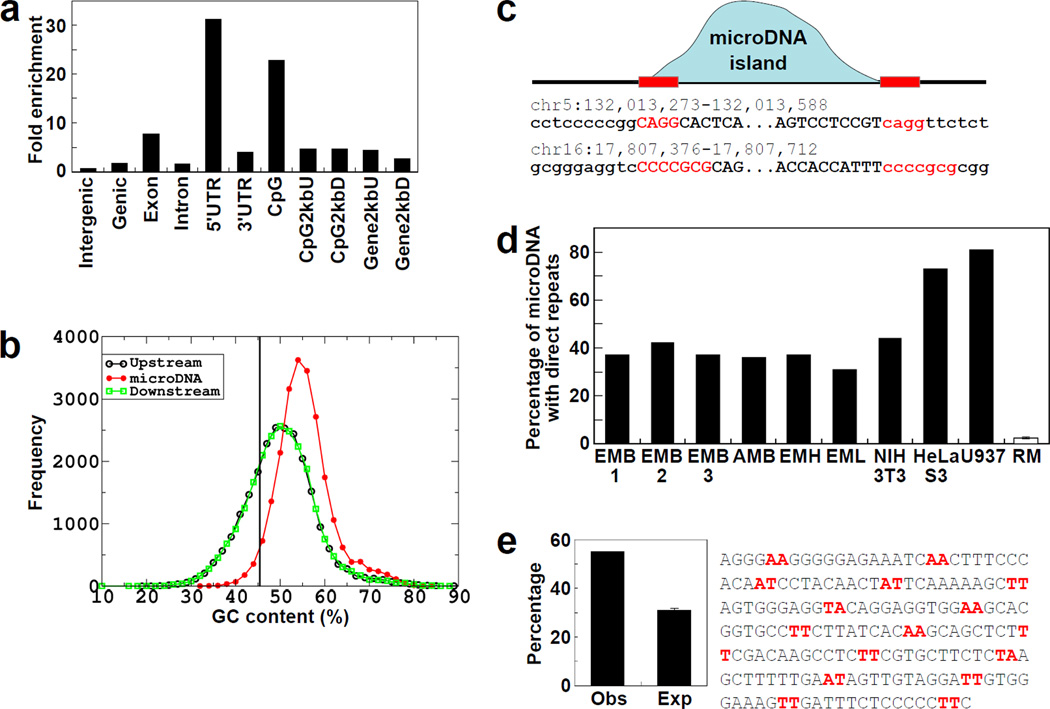
Fig. 2. Properties of the loci that give rise to microDNAs.
a. Enrichment of microDNAs observed in the indicated genomic region relative to the expected percentage based on random distribution. b. Distribution of GC composition in microDNAs in the EMB1 library and their up- and down-stream regions (of same length as microDNA). Vertical line: the genomic average GC content. c. Presence of micro homology near the start and end of a microDNA. "MicroDNA island (blue curve)" is a contiguous stretch of the genome to which the PE-tags map uniquely and correctly. Direct repeats of 2–15 bp (red letters) were observed at the junction of the circle (Upper case) with flanking genomic DNA (Lower case). d. Direct repeats are enriched in different microDNA libraries compared to the random model (RM), generated from the EMB1 sequences. e. Intersection of microDNAs from EMB1 with positioned nucleosome-occupied regions in the mouse liver (13). Obs: observed overlap with nucleosome-occupied DNA. Exp: expected overlap of 1000 randomizations of each microDNA in the library (p<0.0001). A similar enrichment is seen with other microDNA libraries (Fig. S10).