Abstract
Objective
To enhance the pigment production by Streptomyces sp. PM4 for evaluating its anticancer activity.
Methods
Response surface methodology was employed to enhance the production of red pigment from Streptomyces sp. PM4. Optimized pigment was purified and evaluated for the anticancer activity against HT1080, Hep2, HeLa and MCF7 cell lines by MTT assay.
Results
Based on the response surface methodology, it could be concluded that maltose (4.06 g), peptone (7.34 g), yeast extract (4.34 g) and tyrosine (2.89 g) were required for the maximum production of pigment (1.68 g/L) by the Streptomyces sp. PM4. Optimization of the medium with the above tested features increased the pigment yield by 4.6 fold. Pigment showed the potential anticancer activity against HT1080, HEp-2, HeLa and MCF-7cell lines with the IC50 value of 18.5, 15.3, 9.6 and 8.5 respectively.
Conclusions
The study revealed that the maximum amount of pigment could be produced to treat cancer.
Keywords: Streptomyces, Pigment, Response surface methodology, Anticancer activity
1. Introduction
Actinobacteria have been looked upon as potential sources of bioactive compounds and the works done earlier have shown that these microbes are the richest sources of secondary metabolites. They hold a prominent position as targets in screening programs due to their diversity and their proven ability to produce novel metabolites and other molecules of pharmaceutical importance[1]. Since the discovery of actinomycin, actinobacteria have been found to produce many commercially bioactive compounds and natural pigments, in addition to enzymes of industrial importance[2].
The search for an effective anticancer drug to treat cancer has also prompted researchers to investigate the efficacy of natural products in the treatment of cancer. In this respect, actinobacteria are economically and biotechnologically most valuable prokaryotes. They are responsible for the production of about a half of the discovered bioactive secondary metabolites, notably antibiotics, anticancer compounds and enzymes[3]. The vast majority of these compounds are derived from the single actinobacterial genus Streptomyces. Marine Streptomyces have also led to the discovery of a wide range of pigmented cytotoxic compounds with different potency and selectivity from different marine environments. For example, streptochlorin, a yellowish crystalline solid isolated from marine Streptomyces sp. 04DH110[4], blue and red colored ammosamides A and B isolated from the Streptomyces strain CNR-698[5], yellow colored N-carboxamido-staurosporine isolated from marine Streptomyces sp.[6] were have a pronounced anticancer activity. Therefore, marine actinobacteria have attracted serious attention towards the discovery of novel antitumor compounds.
To meet the growing demands in the pharmaceutical industry it is necessary to improve the performance of the system and thus increase the yield. The growth and natural product production of an organism are strongly influenced by medium composition, thus optimization of media components and cultural parameters is the primary task in a biological process[7]. Conventional practice, which allows one variable at a time, does not allow evaluation of the combined effects of all the factors involved in the process and it is a time consuming one. These restrictions can be overcome by the use of Response Surface Methodology (RSM) which can identify and quantify the various interactions among different parameters[8]. In fact, Plackett-Burman design was applied in the first step of optimization to determine the likely effects of medium components on pigment production. The factors having the significant effects were optimized in the second step using a central composite design and the response of each variable was recognized by the regression analysis. Based on the regression analysis, media were optimized for the large scale production[9]. In this study, we report the production of very high levels of a novel red pigment by a Streptomyces sp. PM4 with their potential anticancer activity.
2. Materials and methods
2.1. Actinobacterial strain and its maintenance
The actinobacterial strain PM4 used in this study was isolated from the coral reef environment of Poomarichan Island (lat. 9°14′37.35″N, long. 79°10′58.94″E), Gulf of Mannar Biosphere Reserve, India and it was identified as Streptomyces sp. in accordance with molecular, cultural, morphological and physiological methods. The strain Streptomyces sp. PM4 (GenBank accession: JF445297) was able to produce the red colour reverse side pigment and it was identified as novel based on GC MS analysis. It was maintained on ISP2 agar slants at 4 °C[10].
2.2. Identifying the significant variables using Plackett-Burman Design
In the present study, preliminary investigation of factors affecting the pigment production in the Streptomyces sp. PM4 was done by using Plackett-Burman design. The variables chosen for the present study were investigated at two widely spaced intervals designated as -1 (low level) and +1 (high level). The experimental design for screening the medium components is shown in Table 1 was prepared in seawater. The effects of individual parameters on pigment production were calculated by the following equation:
 |
(1) |
Table 1. Screening for factors affecting the production of pigment by the Streptomyces sp. PM4 using the Plackett-Burman design (g/L).
| Run Order | Glucose | Maltose | Glycerol | Ammonium sulphate | Peptone | Yeast extract | Malt extract | Tyrosine | Phenyl alanine | Histidine | Na2HPo4 | Nacl | Observed Pigment Yield | Predicted Pigment Yield |
| 1 | 1 | 1 | 1 | 0 | 2 | 1 | 2 | 0 | 5 | 5 | 1.5 | 10 | 0.0000 | -0.0000 |
| 2 | 5 | 1 | 1 | 0 | 10 | 5 | 10 | 0 | 0 | 0 | 0.5 | 0 | 0.0652 | 0.0652 |
| 3 | 1 | 5 | 1 | 0 | 10 | 5 | 2 | 5 | 0 | 0 | 1.5 | 10 | 0.3418 | 0.3418 |
| 4 | 5 | 5 | 1 | 0 | 2 | 1 | 10 | 5 | 5 | 5 | 0.5 | 0 | 0.0000 | -0.0000 |
| 5 | 1 | 1 | 5 | 0 | 10 | 1 | 10 | 5 | 0 | 5 | 0.5 | 10 | 0.0000 | 0.0000 |
| 6 | 5 | 1 | 5 | 0 | 2 | 5 | 2 | 5 | 5 | 0 | 1.5 | 0 | 0.0166 | 0.0166 |
| 7 | 1 | 5 | 5 | 0 | 2 | 5 | 10 | 0 | 5 | 0 | 0.5 | 10 | 0.0420 | 0.0420 |
| 8 | 5 | 5 | 5 | 0 | 10 | 1 | 2 | 0 | 0 | 5 | 1.5 | 0 | 0.0282 | 0.0282 |
| 9 | 1 | 1 | 1 | 2 | 2 | 5 | 10 | 5 | 0 | 5 | 1.5 | 0 | 0.0080 | 0.0080 |
| 10 | 5 | 1 | 1 | 2 | 10 | 1 | 2 | 5 | 5 | 0 | 0.5 | 10 | 0.0070 | 0.0070 |
| 11 | 1 | 5 | 1 | 2 | 10 | 1 | 10 | 0 | 5 | 0 | 1.5 | 0 | 0.0000 | 0.0000 |
| 12 | 5 | 5 | 1 | 2 | 2 | 5 | 2 | 0 | 0 | 5 | 0.5 | 10 | 0.0000 | -0.0000 |
| 13 | 1 | 1 | 5 | 2 | 10 | 5 | 2 | 0 | 5 | 5 | 0.5 | 0 | 0.0000 | 0.0000 |
| 14 | 5 | 1 | 5 | 2 | 2 | 1 | 10 | 0 | 0 | 0 | 1.5 | 10 | 0.0000 | 0.0000 |
| 15 | 1 | 5 | 5 | 2 | 2 | 1 | 2 | 5 | 0 | 0 | 0.5 | 0 | 0.0000 | -0.0000 |
| 16 | 5 | 5 | 5 | 2 | 10 | 5 | 10 | 5 | 5 | 5 | 1.5 | 10 | 0.0078 | 0.0078 |
Where, E is the effect of parameter under study and M+ and M- are responses (pigment production) of trials at which the parameter was at its higher and lower levels respectively and N is the total number of trials.
2.3. Optimization by Central Composite Design
Levels of the significant parameters and the interaction effects between various media constituents which influence the pigment production significantly were analyzed and optimized by the Central Composite Design (CCD). In this study, the experimental plan consisted of 31 trials and the independent variables were studied at five different levels (-α, -1, 0, 1, α). All the experiments were done in duplicate and the average of pigment production obtained was taken as the response (Y). The second order polynomial coefficients were calculated and analysed using the Minitab package version 15. The general form of the second degree polynomial equation is:
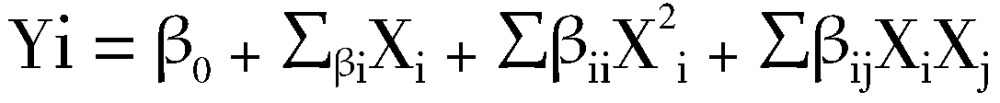 |
(2) |
Where, Yi is the predicted response, XiXj are input variables which influence the response variable Y; β0 is the offset term; βi is the ith linear coefficient; βii is the ith quadratic coefficient and βij is the ijth interaction coefficient.
Statistical analysis of the model was performed to evaluate the analysis of variance (ANOVA). This analysis included the Fisher's F-test (overall model significance), its associated probability P (F), correlation coefficient R and determination coefficient R2 which measure the goodness of fit of regression model. For each variable, the quadratic models were represented as counter plots (3D) and response surface curves were generated using Minitab package version 15.
2.4. Extraction and purification of pigment
Streptomyces sp. PM4 grown in broth culture was centrifuged at 10 000 r/min for 15 min and cell pellet was extracted with methanol and the extraction was centrifuged at 10 000 r/min for 15 min and the white pellet was discarded. Methanol fraction of the pigment was evaporated under reduced pressure and re-extracted with chloroform. Absorbance of the chloroform fraction was measured at 538 nm[10] and estimated with standard plot.
Ten gram of methanol fraction of pigment was passed through the hexane balanced silica gel column (60-120 mesh) to trap the target product within the column. The loaded column was eluted with hexane and ethyl acetate (2:1 v/v) to liberate the adsorbed product. The orange elute was collected and evaporated under reduce pressure to obtain the purified product (red powder).
2.5. Anticancer activity of pigment
The cancer cell lines such as HT1080, Hep2, HeLa and MCF7 were obtained from National Centre for Cell Science, Pune, India. The cell lines were grown to confluence in Dulbecco's Modified Eagle's Medium (DMEM) with 10% FCS. Anticancer effect of pigment was measured by the MTT assay[11]. Cells were seeded on 96-well tissue culture plate with 2×104 cells in 100 µl media/well. After 24 hrs of seeding, cells were cultured in microtiter 96-well plates and treated with different concentrations of pigment for 24 h in a humidified 5% (v/v) CO2/air environment at 37 °C. Subsequently, 20 µl (5 mg/mL) of MTT dye solution was added to each well for 4 h at 37 °C. After the removal of the MTT solution, cells were treated with 150 µl DMSO and the absorbance at 590 nm was quantified using a microplate reader. Decreased cell viability upon pigment treatment was calculated. IC50 value was extrapolated from the plot of viability vs. concentration.
3. Results
3.1. Screening of significant variables using Plackett-Burman Design
This design was applied with 12 different factors as shown in Table 1. All the experiments were performed in duplicate and the average of results (pigment production) were presented as the response pigment yield in Table 1. The main effect of each variable on pigment production was estimated for each independent variable as shown in Table 2.
Table 2. Results of screening experiments using Plackett-Burman design.
| Term | Effect | Coefficient |
| Constant | - | 0.001614 |
| Glucose | -0.001669 | -0.000834 |
| Maltose | 0.002019 | 0.001009 |
| Glycerol | -0.002046 | -0.001023 |
| Ammonium sulphate | -0.002944 | -0.001472 |
| Peptone | 0.002396 | 0.001198 |
| Yeast extract | 0.002789 | 0.001394 |
| Malt extract | -0.001691 | -0.000846 |
| Tyrosine | 0.001536 | 0.000768 |
| Phenyl alanine | -0.002311 | -0.001156 |
| Histidine | -0.002679 | -0.001339 |
| Na2HPo4 | 0.001801 | 0.000901 |
| Nacl | 0.001754 | 0.000877 |
Results have indicated that the presence of higher levels of maltose, peptone, yeast extract and tyrosine in the culture medium was affected the pigment production positively. On the other hand, NH2HPO4 and NaCl showed little effect on pigment production. Glucose, glycerol, ammonium sulphate, malt extract, phenyl alanine and histidine showed negative effect on pigment production.
3.2. Optimization of pigment production using Central Composite Design
Preliminary experiments showed that maltose, peptone, yeast extract and tyrosine influenced the pigment production effectively. So, Central Composite Design was employed to determine the optimum level of these parameters leading to maximum pigment synthesis. Level of these factors used in optimization studies was determined by Plackett-Burman Design. The effect of the four variables, each at five levels and their interactions on pigment production were determined by carrying out 31 experiments and the results are presented in Table 3.
Table 3. Experimental design and results along with the observed and predicted values of pigment for CCD of response surface methodology (g/L).
| Run order | Maltose | Peptone | Yeast extract | Tyrosine | Observed Pigment Yield | Predicted Pigment Yield |
| 1 | 3 | 5 | 3 | 2 | 0.398 | 0.420075 |
| 2 | 5 | 5 | 3 | 2 | 0.940 | 0.892900 |
| 3 | 3 | 10 | 3 | 2 | 0.212 | 0.192233 |
| 4 | 5 | 10 | 3 | 2 | 0.320 | 0.354108 |
| 5 | 3 | 5 | 6 | 2 | 0.172 | 0.127300 |
| 6 | 5 | 5 | 6 | 2 | 0.422 | 0.458075 |
| 7 | 3 | 10 | 6 | 2 | 0.186 | 0.177408 |
| 8 | 5 | 10 | 6 | 2 | 0.242 | 0.197233 |
| 9 | 3 | 5 | 3 | 4 | 0.196 | 0.190233 |
| 10 | 5 | 5 | 3 | 4 | 0.248 | 0.293108 |
| 11 | 3 | 10 | 3 | 4 | 0.322 | 0.322442 |
| 12 | 5 | 10 | 3 | 4 | 0.1202 | 0.114367 |
| 13 | 3 | 5 | 6 | 4 | 0.136 | 0.138408 |
| 14 | 5 | 5 | 6 | 4 | 0.130 | 0.099233 |
| 15 | 3 | 10 | 6 | 4 | 0.552 | 0.548567 |
| 16 | 5 | 10 | 6 | 4 | 0.184 | 0.198442 |
| 17 | 2 | 7.5 | 4.5 | 3 | 0.246 | 0.267658 |
| 18 | 6 | 7.5 | 4.5 | 3 | 0.398 | 0.390358 |
| 19 | 4 | 2.5 | 4.5 | 3 | 0.384 | 0.388325 |
| 20 | 4 | 12.5 | 4.5 | 3 | 0.250 | 0.259692 |
| 21 | 4 | 7.5 | 1.5 | 3 | 0.386 | 0.367358 |
| 22 | 4 | 7.5 | 7.5 | 3 | 0.126 | 0.158658 |
| 23 | 4 | 7.5 | 4.5 | 1 | 0.35 | 0.379325 |
| 24 | 4 | 7.5 | 4.5 | 5 | 0.166 | 0.150692 |
| 25 | 4 | 7.5 | 4.5 | 3 | 1.618 | 1.676857 |
| 26 | 4 | 7.5 | 4.5 | 3 | 1.680 | 1.676857 |
| 27 | 4 | 7.5 | 4.5 | 3 | 1.700 | 1.676857 |
| 28 | 4 | 7.5 | 4.5 | 3 | 1.720 | 1.676857 |
| 29 | 4 | 7.5 | 4.5 | 3 | 1.620 | 1.676857 |
| 30 | 4 | 7.5 | 4.5 | 3 | 1.700 | 1.676857 |
| 31 | 4 | 7.5 | 4.5 | 3 | 1.700 | 1.676857 |
Analysis of variance (ANOVA) for the concentration of pigment is depicted in Table 4. This gives the value of the model and determines the requirement of a more complex model with a better fit. The value of R2 was 0.9976, indicating that the model was fitted and explains 99.76% of variability in pigment concentration. F-Test for the regression was significant at the level of 5% (P<0.05), indicating that the model is fit and can adequately explain the variations observed in pigment concentration with the designed level of factors. If the F-test for lack of fit is significant, then a more complicated model is needed to fit the data. As seen in Table 4, the lack of fit (0.553) was not significant at the 5% level (P<0.05), indicating that the experimental data obtained fitted well with the model. These results show that the model chosen can satisfactorily explain the effects of maltose, peptone, yeast extract and tyrosine on pigment production in submerged cultures. The following second order polynomial equation was found to explain pigment production by the Streptomyces sp. PM4.
Table 4. Analysis of variance for the fitted quadratic polynomial model of pigment production.
| Source | DF | Seq SS | Adj SS | Adj MS | F | P |
| Regression | 14 | 11.0622 | 11.0622 | 0.79016 | 481.89 | 0.000 |
| Linear | 4 | 0.1911 | 3.6861 | 0.92153 | 562.00 | 0.000 |
| Square | 4 | 10.3524 | 10.3524 | 2.58810 | 1578.38 | 0.000 |
| Interaction | 6 | 0.5187 | 0.5187 | 0.08645 | 52.72 | 0.000 |
| Residual Error | 16 | 0.0262 | 0.0262 | 0.00164 | - | - |
| Lack-of-Fit | 10 | 0.0161 | 0.0161 | 0.00161 | 0.95 | 0.553 |
| Pure Error | 6 | 0.0102 | 0.0102 | 0.00170 | - | - |
| Total | 30 | 11.0885 | - | - | - | - |
Y=-0.72067+1.58304X1+0.46137X2+0.68053X3+1.08541X4 -1.73534X12-0.29085X22-0.81571X32-1.83534X42-0.12275X1X2 -0.06375X1X3-0.22438X1X4+0.02950X2X3+ 0.04775X2X4+ 0.14708X3X4; Where Y is the pigment production and X1, X2, X3 and X4 are the coded values of the test variables (maltose, peptone, yeast extract and tyrosine).
Regression analysis of the experimental data showed that maltose, peptone, yeast extract and tyrosine had positive linear effects on pigment production (P<0.05) when the probability (P-) values were used as a tool to check the significance of each of the co-efficients. The more significant one was the correlation with the corresponding co-efficient (Table 5). Among the four factors tested, maltose (1.58304) had the highest impact on pigment concentration, as given by the linear co-efficient followed by tyrosine (1.08541), yeast extract (0.68053) and peptone (0.46137). These factors also showed significant negative quadratic effects on pigment production, indicating that pigment concentration, increased and decreased as its level increased above certain value. Interaction between these parameters was also significant, as shown by low P-values (P<0.05) for interactive terms.
Table 5. Regression coefficients and their significance of the quadratic model of pigment production.
| Term | Coef | SE Coef | T | P |
| Constant | -0.72067 | 0.014734 | -48.911 | 0.000 |
| Maltose | 1.58304 | 0.036822 | 42.992 | 0.000 |
| Peptone | 0.46137 | 0.013686 | 33.710 | 0.000 |
| Yeast extract | 0.68053 | 0.022811 | 29.834 | 0.000 |
| Tyrosine | 1.08541 | 0.034216 | 31.723 | 0.000 |
| Maltose*Maltose | -1.73534 | 0.034571 | -50.197 | 0.000 |
| Peptone*Peptone | -0.29085 | 0.005531 | -52.583 | 0.000 |
| Yeast extract*Yeast extract | -0.81571 | 0.015365 | -53.089 | 0.000 |
| Tyrosine*Tyrosine | -1.83534 | 0.034571 | -53.089 | 0.000 |
| Maltose*Peptone | -0.12275 | 0.018487 | -6.640 | 0.000 |
| Maltose*Yeast extract | -0.06375 | 0.030811 | -2.069 | 0.050 |
| Maltose*Tyrosine | -0.22438 | 0.046217 | -4.855 | 0.000 |
| Peptone*Yeast extract | 0.02950 | 0.012324 | 2.394 | 0.029 |
| Peptone*Tyrosine | 0.04775 | 0.018487 | 2.583 | 0.020 |
| Yeast extract*Tyrosine | 0.14708 | 0.030811 | 4.774 | 0.000 |
Fitted response for the above regression model was plotted (Figure 1). The graphs were generated for the pair-wise combination of the 2 factors while keeping the other middle level for the pigment production. The main goal of response surface is to efficiently hunt for the optimum values of the variables, such that the response is maximized. The surface plots affirm that the objective function is unimodal in nature, which shows an optimum in the centre. The central optimum point was evaluated using gradient method in the direction of steepest ascend of media for the pigment production evaluated from the surface plots. The optimal values of maltose, peptone, yeast extract and tyrosine were estimated in actual units and they were 4.06 g, 7.34 g, 4.34 g and 2.89 g, respectively, with a predicted pigment concentration of 1.684 g/L in submerged fermentation. Confirmation experiment was conducted for these predicted optimum conditions and pigment concentration from the experiment was observed as 1.874 g/L. This was higher than the predicted value which reveals the higher accuracy of the model.
Figure 1. Statistical optimization of the pigment production using RSM. (A) Yeast extract and tyrosine; (B) maltose and peptone; (C) maltose and tyrosine; (D) peptone and yeast extract; (E) maltose and yeast extract; (F) peptone and tyrosine.
3.3. Anticancer activity of pigment
Anticancer activity of the purified pigment against HT1080, HEp-2, HeLa and MCF-7 cell line was showed in figure 2. The pigment was found to be effective against all the four cancer cell lines. The IC50 value of the pigment was 18.5, 15.3, 9.6 and 8.5 against HT1080, HEp-2, HeLa and MCF-7cell lines respectively.
Figure 2. Inhibition of cell viability by pigment treatment under in vitro conditions.
4. Discussion
While there are several reports about the control of secondary metabolite production by actinobacteria[12],[13], optimization of cultivation conditions is expected to improve the secondary metabolite production. In this study, optimization of pigment production by the strain of Streptomyces sp. PM4 using two different statistical designs was investigated.
Major approaches used for screening and media optimization have been reported by Parekh et al[14]. However, one of the basic approaches used to design experiments is to screen media components based on empirical processes. Statistical techniques for experimental design provide with a more accurate and elegant means of designing the best medium[15]. The most widely used statistical experimental designs are Plackett-Burman design and Central Composite design[16]. Advantages of these designs include simplicity and assessment of a large number of factors on the relative efficiency of the production process.
The Plackett-Burman design does not yield estimates of the extent or type of interaction between variables[17]. Tweleve factors were investigated to determine the optimum medium components suitable for pigment production by the Streptomyces sp. PM4. The statistical analysis of the activities obtained from the 12 experiments revealed that maltose, peptone, yeast extract and tyrosine are the significant factors influencing the production of pigment.
Taking into consideration the basic role of carbon source in enhancing pigment production, two justifications can be made. The first point is that in the medium, which is basically devoid of carbon source, the addition of maltose enhanced the pigment production. The second point is that the decrease in pigment production seen with the addition of glycerol and glucose as a sole carbon sources, could be due to a catabolite repression[18].
In the present study, the Streptomyces sp. PM4 significantly utilized the yeast extract and peptone. Yeast extract contains the simpler compounds such as amino acids and peptides with the rich sources of vitamins especially those belonging to B- complex. Peptones also contain small amounts of various organic and inorganic compounds. But they may be deficient in certain minerals and vitamins. Both peptone and yeast extract were significantly increase the pigment production by providing the suitable nutrients for the growth and pigment production of the Streptomyces sp. PM4. Amino acids play a crucial role in the pigment production by the formation of the precursor molecule. In the present study, amino acid, tyrosine enhanced the pigment production, indicating that it is responsible for the enhancement of pigment production by incorporating into pigment structure. RSM used in this investigation suggested the importance of various fermentation parameters at different levels. Based on the results of screening experiments by Plackett-Burman design, variables such as maltose, peptone, yeast extract and tyrosine were selected and further optimized using Central Composite Design. This revealed a high similarity between the predicted and experimental results, which reflected the accuracy and applicability of RSM to optimize the process for pigment production in submerged culture.
Effect of the interaction of various chemical parameters on pigment production by the Streptomyces sp. PM4 was investigated by plotting the response surface curves against any two independent variables while keeping the third independent variable at the ‘0’ level. Thus 6 response surfaces were obtained by considering all the possible combinations. Growth of the organism and pigment production increased with the increase in all the parameters up to the optimum level, after which growth declined and the pigment production also dropped. This could be due to the loss of nutrient in culture medium and due to the prolonged incubation.
From the response surface graphs, it could be concluded that the optimum values of maltose, peptone, yeast extract and tyrosine for the maximum production of pigment (1.68 g/L) by the Streptomyces sp. PM4 were 4.06 g, 7.34 g, 4.34 g and 2.89 g, respectively. In order to validate the adequacy of the model, verification experiments were conducted under predicted optimal fermentation conditions. The results have indicated that the model is satisfactory.
Progress has been made recently on drug discovery from actinobacteria by using high-throughput screening and fermentation, mining genomes for cryptic pathways, and combinatorial biosynthesis to generate new secondary metabolites related to existing pharmacophores[13]. In the present study, anticancer effect of the actinobacterial pigment on cervical (HeLa), Larynx (HEp-2), fibro sarcoma (HT1080) and breast (MCF-7) cancer cell line has been studied. Pigment showed cytotoxic in a dose-dependent manner. Maximum cytoxicity of the pigment was observed at 24 h incubation in a concentration dependent manner. It was found that 50 µg/mL of pigment could greatly inhibit the cell growth. The IC50 value of 18.5, 15.3, 9.6 and 8.5 against HT1080, HEp-2, HeLa and MCF-7 cell lines respectively. Similarly, the polyketide yellow pigment from Streptomyces sp. CNQ766 having cytotoxicity against Mouse splenocyte T-cells and macrophages with the IC50 value of 20 µg/mL[19]. Ravikumar et al.[20] also reported the anticancer property of sediment actinomycetes against MCF-7 and MDA-MB-231 cell lines with the IC50 value range from 10.13 to 18.54. Thus the present study also indicating that the pigment is having potential cytotoxic activity against cancer cell lines.
Currently, there is a lot of interest in the scientific community around the world in exploiting novel anticancer drugs. Marine actinobacteria, with their unique nature, differ very much in many aspects from their terrestrial counterparts and are known to produce diverse spectra of novel useful substances. In this context, results obtained in the present study indicate the scope for utilizing the pigment produced from Streptomyces sp. PM4 for the development of anticancer drugs.
Acknowledgments
Authors thank the Director and Dean of the Centre of Advanced Study in Marine Biology and the Authorities of Annamalai University for providing with necessary facilities. Authors also thank Ministry of Environment and forests, India for financial assistance (Grant No. MoEn&F/22/27/2005-CSC).
Comments
Background
The search for an effective anticancer drug to treat cancer has also prompted researchers to investigate the efficacy of natural products in the treatment of cancer. In this respect, actinobacteria having great potential to produce drugs effectively against cancer economically and biotechnologically most valuable.
Research frontiers
SRSM used in this investigation suggested the importance of various fermentation parameters at different levels. Plackett-Burman design and Central Composite Design was used to optimize effective medium composition with different levels. The study revealed that the maximum amount of pigment could be produced to treat cancer.
Related reports
Based upon the reference used in this study reveals effective drugs produced by the actinobacteria but according to this study the methods were used to standardize the medium to produce large quantity by using response surface designing of sources of medium.
Innovations and breakthroughs
The methods were followed in this study are very innovative and predict the effective medium concentration with standard statistical methods. The most effective and potential compound against cancer producing by actinobacteria shows significant property against cancer.
Applications
The Streptomyces sp. PM4 was identified and used for producing drug anticancer. The study concluded that the potential bacteria and its red pigment was effectively controlled the cancer. Furthermore study needs to confirm the potential of the compound with animal studies.
Peer review
This is the need of hour in producing effective drug against anticancer drugs. The marine actinobacteria Streptomyces sp. PM4 is important candidates to treat various dreadful diseases. Response surface methodology was employed in this study was successful to standardize the requirements of medium. The cell lines were used for anticancer activity and pigment showed the potential anticancer activity against HT1080, HEp-2, HeLa and MCF-7 cell lines with the IC50 value of 18.5, 15.3, 9.6 and 8.5 respectively. This is very significant activity showed by Streptomyces sp. PM4.
Footnotes
Foundation Project: Financially supported by Ministry of Environment and Forests, India (Grant No. MoEn&F/22/27/2005-CSC).
Conflict of interest statement: We declare that we have no conflict of interest.
References
- 1.Balachandran C, Duraipandiyan V, Ignacimuthu S. Purification and characterization of protease enzyme from actinomycetes and its cytotoxic effect on cancer cell line (A549) Asian Pac J Trop Biomed. 2012;2(Suppl 2):S1138–S1146. doi: 10.1016/S2221-1691(12)60215-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saranya S, Edith Rena A, Karuppiah V, Sivakumar K. Actinobacteria of the Bitharkanika mangroves of India: Sreening for antibiosis and industrially important enzymes. Natl Acad Sci Lett. 2011;34:393–400. [Google Scholar]
- 3.Manivasagan P, Venkatesan J, Sivakumar K, Kim SK. Marine actinobacterial metabolites: Current status and future perspectives. Microbiol Res. 2013 doi: 10.1016/j.micres.2013.02.002. http://dx.doi.org/10.1016/j.micres.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Shin JH, Jeong HS, Lee HS, Park SK, Kim HM, Kwon HJ. Isolation and structure determination of streptochlorin, an antiproliferative agent from a marine-derived Streptomyces sp. 04DH110. J Microbiol Biotechnol. 2007;17(8):1403–1406. [PubMed] [Google Scholar]
- 5.Hughes CC, MacMillan JB, Gaudêncio SP, Jensen PR, Fenical W. The ammosamides: structures of cell cycle modulators from a marine-derived Streptomyces species. Angew Chem Int Ed Engl. 2009;48(4):725–727. doi: 10.1002/anie.200804890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu SJ, Fotso S, Li F, Qin S, Kelter G, Fiebig HH, et al. et al. N-carboxamido-staurosporine and selina-4(14), 7(11)-diene- 8,9-diol, new metabolites from a marine Streptomyces sp. J Antibiot. 2006;59(6):331–337. doi: 10.1038/ja.2006.46. [DOI] [PubMed] [Google Scholar]
- 7.Usha R, Mala KK, Venil CK, Palaniswamy M. Screening of Actinomycetes from mangrove ecosystem for L-asparaginase activity and optimization by response surface methodology. Polish J Microbiol. 2011;60(3):213–221. [PubMed] [Google Scholar]
- 8.Sivakumar K, Karuppiah V, Vijayabaskara Sethubathi G, Thangaradjou T, Kannan L. Response surface methodology for the optimization of amylase production by Streptomyces sp. ML12 using agricultural byproducts. Biologia. 2012;67(1):32–40. [Google Scholar]
- 9.Zhao W, Zheng J, Wang YG, Zhou HB. A marked enhancement in production of amylase by Bacillus amyloliquefaciens in flask fermentation using statistical methods. J Cent South Univ T. 2011;18(4):1054–1062. [Google Scholar]
- 10.Karuppiah V. Ecology, diversity and bioactivity of marine actinobacteria of the gulf of mannar biosphere reserve. 2011. India. Ph.D. Thesis, Annamalai University.
- 11.Rajendra Prasad N, Karthikeyan A, Karthikeyan S, Venkata Reddy B. Inhibitory effect of caffeic acid on cancer cell proliferation by oxidative mechanism in human HT-1080 fibrosarcoma cell line. Mol Cell Biochem. 2011;349:11–19. doi: 10.1007/s11010-010-0655-7. [DOI] [PubMed] [Google Scholar]
- 12.Sivakumar K, Sahu MK, Thangaradjou T, Kannan L. Research on marine actinobacteria in India. Indian J Microbiol. 2007;47:186–196. doi: 10.1007/s12088-007-0039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olano C, Mendez C, Salas JA. Antitumor compounds from actinomycetes: from gene clusters to new derivatives by combinatorial biosynthesis. Nat Prod Rep. 2009;26(5):628–660. doi: 10.1039/b822528a. [DOI] [PubMed] [Google Scholar]
- 14.Parekh S, Vinei VA, Strobel RJ. Improvement of microbial strains and fermentation process. Appl Microbiol Biotechnol. 2000;54:287–301. doi: 10.1007/s002530000403. [DOI] [PubMed] [Google Scholar]
- 15.Thenmozhi S, Sankar R, Karuppiah V, Sampath Kumar P. L-asparaginase production by mangrove derived Bacillus cereus MAB5: Optimization by response surface methodology. Asian Pac J Trop Med. 2011;4(6):486–491. doi: 10.1016/S1995-7645(11)60132-6. [DOI] [PubMed] [Google Scholar]
- 16.Prajapati VS, Trivedi UB, Patel KC. Optimization of glucoamylase production by Colletotrichum sp. KCP1 using statistical methodology. Food Sci Biotechnol. 2013;22(1):31–38. [Google Scholar]
- 17.Patil SA, Surwase SN, Jadhav SB, Jadhav JP. Optimization of medium using response surface methodology for l-DOPA production by Pseudomonas sp. SSA. Biochem Eng J. 2013;74:36–45. [Google Scholar]
- 18.Giri VA, Anandkumar N, Muthukumaran G, Pennathur G. A novel medium for enhanced cell growth and production of prodigiosin from Serratia marcescens isolated from soil. BMC Microbiol. 2004;4:1–10. doi: 10.1186/1471-2180-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho JY, Kwon HC, Williams PG, Kauffman CA, Jensen PR, Fenical W. Actinofuranones A and B, polyketides from a marine-derived bacterium related to the genus Streptomyces (Actinomycetales) J Nat Prod. 2006;69(3):425–428. doi: 10.1021/np050402q. [DOI] [PubMed] [Google Scholar]
- 20.Ravikumar S, Fredimoses M, Gnanadesigan M. Anticancer property of sediment actinomycetes against MCF-7 and MDA-MB-231 cell lines. Asian Pac J Trop Biomed. 2012;2:92–96. doi: 10.1016/S2221-1691(11)60199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]




