Abstract
This is the first report of imaging mass spectrometry (MS) from multiply charged ions at vacuum. Laserspray ionization (LSI) was recently extended to applications at vacuum producing electrospray ionization-like multiply charged ions directly from surfaces using a commercial intermediate pressure matrix-assisted laser desorption/ionization ion mobility spectrometry (IMS) MS instrument. Here, we developed a strategy to image multiply charged peptide ions. This is achieved by the use of 2-nitrophloroglucinol as matrix for spray deposition onto the tissue section and implementation of ‘soft’ acquisition conditions including low laser power and lower ion accelerating voltages similar to electrospray ionization-like conditions. Sufficient ion abundance is generated by the vacuum LSI method to employ IMS separation in imaging multiply charged ions obtained on a commercial mass spectrometer ion source without physical instrument modifications using the laser in the commercially available reflection geometry alignment. IMS gas-phase separation reduces the complexity of the ion signal from the tissue, especially for multiply charged relative to abundant singly charged ions from tissue lipids. We show examples of LSI tissue imaging from charge state +2 of three endogenous peptides consisting of between 1–16 amino acid residues from the acetylated N-terminal end of myelin basic protein: m/z 795.81 (+2) MW 1589.6, m/z 831.35 (+2) MW 1660.7, and m/z 917.40 (+2) MW 1832.8.
Keywords: Laserspray Ionization (LSI), intermediate pressure, vacuum, MALDI, imaging mass spectrometry, ion mobility spectrometry (IMS), myelin basic protein (MBP), peptide, multiply charged, 2-nitrophloroglucinol (2-NPG) matrix
Introduction
Matrix-assisted laser desorption/ionization (MALDI) mass spectrometry (MS) produces abundant singly charged ions of peptides from the solid state under low pressure, intermediate pressure, and atmospheric pressure conditions.1–3 Time-of-flight (TOF) is the common mass analyzer used with low pressure MALDI and in principle provides unlimited mass range for detection of large molecules.4 A limitation of the production of primarily singly charged ions is that high-mass compounds cannot be analyzed using high performance instruments with only a limited mass range such as Fourier transform ion cyclotron resonance, Orbitrap, or ion mobility spectrometry (IMS) (SYNAPT G2) mass spectrometers. MALDI imaging mass spectrometry has provided information on spatial distributions of lipids,5,6 peptides,7 and proteins8 directly from biological tissue sections, and MALDI-IMS-MS imaging has been applied for lipids6,9 and peptides6 as well as studying protein changes related to several diseases such as Alzheimer’s and Parkinsons10–12 as well as cancer.13–15 Secondary ion mass spectrometry (SIMS) has also been used to map the distribution of cancer chemotherapeutic agents in tissues and cells,16,17 lipids,18–20 and later, matrix-enhanced SIMS to enhanced subcellular imaging of tissue samples.21–23 Desorption electrospray ionization is promising for imaging living tissue without sample preparation.24–26
Laserspray ionization inlet (LSII)27–35 and matrix assisted ionization inlet (MAII)36 are two new ionization methods in MS operating from atmospheric pressure to produce highly charged ions similar to electrospray ionization (ESI). Both LSII and MAII have attributes in common with MALDI allowing analysis of compounds directly from the solid state by using a solid matrix material comprised of a small organic compound.37 The application of heat and the presence of an atmospheric pressure to vacuum pressure drop region are two important factors in producing abundant highly charged ions in the inlet of the mass spectrometer.29,38,39 In LSII, the laser alignment is not important because multiply charged ions are formed without the use of a laser,36,37,40 as well as with the use of a laser aligned in transmission28–33 or reflection geometry.41 For example, a commercial atmospheric pressure MALDI source with the laser aligned in reflection geometry produced abundant highly charged protein (e.g., lysozyme) ions on a Thermo Orbitrap mass spectrometer.41 Critical is that sufficient heat is provided to the inlet of the mass spectrometer which directly depends on the matrix used.29,37 Multiply charged ions produced by LSII and MAII using 2-nitrophologlucinol (2-NPG) matrix, extend the mass range of detection to at least bovine serum albumin (~66 kDa).43 LSII was also shown to produce sufficient ion abundance directly from tissue to identify an endogenous peptide, a fragment of N-acetylated myelin basic protein (MBP) (1–16), using electron transfer dissociation (ETD) and high accuracy mass measurement,31 and has been successfully introduced on a mass spectrometer with a heated inlet to perform imaging of lipids.34,35 The laser aligned in transmission geometry provides ease of operation,27–33 and offers the potential for high spatial resolution imaging.31,34,35
Initial studies on matrixes30 enabled us to introduce LSI technology operating from vacuum, LSIV, using a commercial intermediate pressure MALDI ion source interfaced to a Waters IMS-MS SYNAPT G2 mass spectrometer.42 In the absence of a heated inlet region, abundant multiply charged analyte ions are formed by laser ablation using a matrix that efficiently evaporates.37 2-NPG matrix is the first matrix to produce highly charged ions at atmospheric pressure, intermediate pressure, and low pressure.39,40,43 The mechanism proposed for the LSI inlet and vacuum ionization process is laser ablation producing highly charged matrix/analyte droplets which upon matrix evaporation releases ions by a similar process to that proposed for ESI.44,45 The impact of a laser has been experimentally and theoretically shown to form molten droplets of matrix/analyte and the existence of clusters was verified recently for LSII.38,46,47 2,5-Dihydroxyacetophenone (2,5-DHAP),31,33 a matrix that requires a lower inlet temperature than, for example, 2,5-DHB produces abundant multiply charged ions at intermediate pressure under LSI tuning conditions. 39,42 However, the use of higher laser power42 and voltages39 increase the abundance of singly charged ions. Using a higher laser power makes only minor contribution to the production of singly charged ions. It is primarily the higher voltages of the MALDI settings that allow switching from multiply charged using ESI-like settings to singly charged ions either at atmospheric pressure41 or intermediate pressure39.
The practical utility of LSIV on a commercial vacuum MALDI source in combination with IMS-MS was demonstrated using 2,5-DHAP as matrix for the mixture analysis of lipids, peptides, and proteins.42 Various families of compounds are well separated from each other. Imaging applications are an extension to these tissue analysis developments. Here, we show the applicability of LSIV using a commercially available vacuum MALDI source operating at intermediate pressure (~0.16 Torr) for tissue imaging of multiply charged peptide ions achieved without any physical instrument modifications.
Experimental Section
Materials
α-Cyano-4-hydroxycinnamic acid (CHCA), 2-NPG, and bovine insulin were purchased from Sigma Aldrich Co. (St. Louis, MO), MBP peptide from Anaspec (Fremont, CA), and galanin from America Peptide Co. (Sunnyvale, CA). 2,5-DHAP, acetonitrile (ACN), ethanol (EtOH), and formic acid (FA) were purchased from Fisher Scientific Inc. (Pittsburgh, PA) and acetic acid (AA) from Mallinckdrodt Chemicals (Phillipsburg, NJ). Methanol (MeOH) and HPLC grade water were obtained from EMD Chemicals (Gibbstown, NJ). Microscopy glass plates from Gold Seal (Portsmouth, NH) were used as sample target plates.
Sample Preparation
Stock solutions of synthesized standard N-acetylated MBP peptide and galanin were prepared in water and bovine insulin in 50:50 MeOH:water with 1% AA, and diluted to 1 pmol µL−1 in water. 2,5-DHAP and 2-NPG matrix solutions were prepared by dissolving 5 mg in 150 µL of 50:50 ACN:water (warmed), and 100 and 300 µL of 50:50 ACN:water (with and without 0.1% FA), respectively. The matrix:analyte mixture was prepared in 1:1 volume ratio before depositing 1 µL of the analyte:matrix mixture on a glass plate using the dried droplet method.42 Mouse brain tissue sections were obtained as previously described.9,31,34,35 The tissue sections were mounted on plain and precoated glass plates with CHCA matrix using a tissue box,34 and delipified with EtOH as previously described.31 The delipified tissue slices were spotted separately with several 0.5 µL of 100% 2,5-DHAP and 100% 2-NPG matrix in 50:50 ACN:water for direct analysis and spray coated with 2-NPG for imaging. A binary mixture of 10% 2-NPG and 90% 2,5-DHAP matrix solutions was also used for tissue imaging.
Laserspray Ionization Vacuum (LSIV)
A commercial SYNAPT G2 intermediate pressure MALDI mass spectrometer from Waters Co. (Manchester, England) equipped with an IMS was employed in this study. The intermediate pressure MALDI source operates using a Nd:YAG laser (355 nm) in reflection geometry mode (Scheme 1). The laser operates with an average power of 20 mW and a pulse energy of 100 µJ at 200 Hz firing rate. Once the sample plate was loaded, the source pressure was under intermediate pressure vacuum condition at ~0.2 mbar.42 ‘LSI (ESI-like) settings’ with lower voltages, and commercial ‘MALDI settings’ were employed in this study, as previously described. 39,42 Briefly, acquisitions were performed in positive ion and sensitivity modes using the following settings: 0 V (LSI) and 20 V (MALDI) on sample plate, 10 V “extraction”, 10 V “hexapole bias”, and 5 V “aperture 0”. In the analysis of standards (MBP, galanin, bovine insulin), the laser power used ranged from 4.2 to 7.3 J cm−2 39 at a firing rate of 200 Hz and a scan rate of 1 scan per second acquiring a sum mass spectrum of 1 to 2 min. For tissue imaging, the instrument’s MALDI imaging pattern creator software was used as one would use for MALDI imaging. The sampling resolution was set at 100 µm×100 µm x,y laser step size, similar to MALDI imaging. Acquisition was set at a scan time of 2 seconds, laser power of 12.5 J cm−2 39 at a firing rate of 200 Hz, and 1 scan time per pixel. The total imaging acquisition time was ~5 hrs to image the entire tissue section.
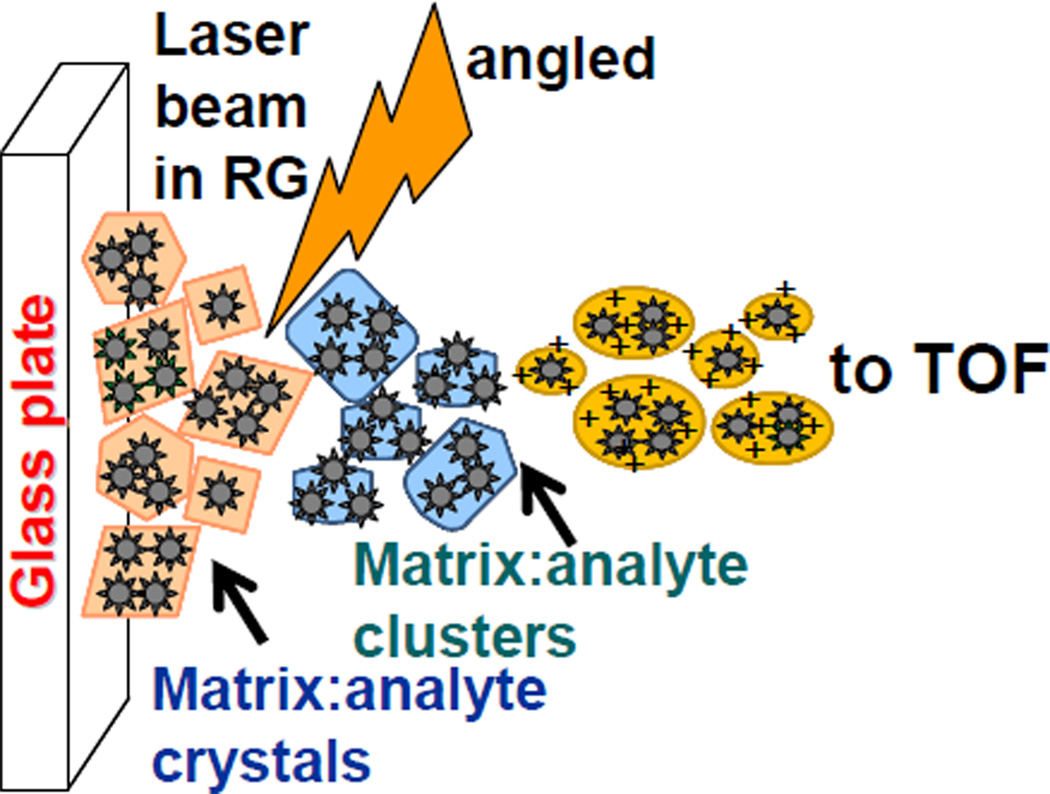
Scheme 1.
Cluster model representation of LSIV at intermediate pressure in reflection geometry laser ablation of the matrix/analyte crystals forming charged matrix/analyte clusters. Related ionization methods are shown in Supplemental Information.
Laserspray Ionization Inlet (LSII)
A Nanolockspray source of the SYNAPT G2 was used to perform LSII at atmospheric pressure as described in previous studies.32,33 A homebuilt skimmer cone was used with a copper inlet tube and the laser was aligned at 180° to the inlet tube, focused onto the matrix:analyte sample on a glass plate, and ablated in transmission geometry (Scheme S1A) relative to the inlet of the mass spectrometer. The source temperature was set at 150 °C and acquisition was obtained in resolution mode using 100 V and 4 V sample cone and extraction cone voltages, respectively, with a scan time of 1 second for 2 min.
Data Processing
MassLynx version 4.1 was used to process the mass spectra, DriftScope version 2.2 was used to extract the 2-dimensional (2-D) plot of drift time vs. mass-to-charge (m/z) ratio, and mobility extractor to obtain the drift time versus m/z selected dataset and convert into an MSI file using the MALDI imaging converter. BioMAP 3.8.0.1 was used from Novartis Institutes for Biomedical Research, Basel, Switzerland, to extract from the MSI file the images of the peptides.
Results and Discussion
The endogenous N-acetylated MBP peptide (Ac-ASQKRPSQRSKYLATA), previously sequenced by LSII (Scheme S1A) using high performance fragmentation,31 was the initial focus for obtaining MS images of multiply charged ions using LSIV (Scheme 1) on a commercial intermediate pressure MALDI source of the SYNAPT G2 mass spectrometer. The use of a suitable matrix material and appropriate settings and acquisition conditions such as laser power and voltages for ion acceleration and transmission are important factors influencing the formation of the highly charge ions at intermediate pressure.37,39,42 The direct analysis of MBP peptide from delipified mouse brain tissue was first achieved by spotting 2,5-DHAP matrix onto the tissue. Multiply charged peptide ions up to charge state +3 were detected with the largest MW of ~3.2 kDa (Figure S1). Exceptional ion abundance is obtained for the MBP peptide with m/z 917.39 using 2,5-DHAP, but only when the matrix is directly spotted on the tissue (Figure S1A). With LSII, spotting 2,5-DHAP matrix on non-delipified mouse brain tissue allowed detection of lipids as well as multiply charged ions of the N-acetylated MBP fragment peptide (Figure S2). However, spray coating of the matrix, necessary for good spatial resolution in tissue imaging, provided poor ion abundance using 2,5-DHAP operated from atmospheric pressure or vacuum.
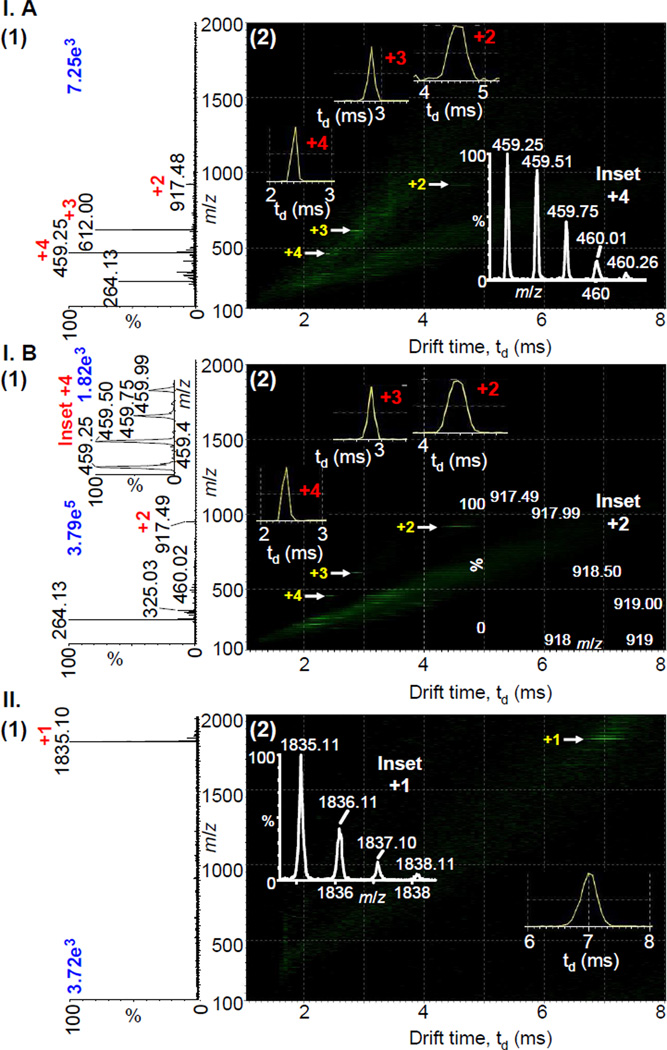
2-NPG, a more volatile matrix than 2,5-DHAP, produced abundant multiply charged ions of up to charge state +4 from a synthesized MBP peptide standard (Figure 1I.A) at low laser power applying ‘LSI settings’39,42 on the commercial intermediate pressure MALDI source without any physical instrument modifications. The analyte ion intensity increased as did the chemical noise background with increasing laser power (Figure 1I.B). The chemical background could be removed using the IMS dimension, especially because the different charge state ions are well separated in the IMS dimension, identical to ESI-IMS-MS.48–53 Only singly charged ions are produced when the default ‘MALDI settings’39 are applied (Figure 1II). The ion abundances observed are similar for the multiply charged ions (‘LSI settings’) (Figure 1I) and the singly charged ion (‘MALDI settings’).39 For a number of peptide and small protein standards using ‘LSI settings’39,42 (Figure S3: (A) MBP, MW 1833 Da; (B) galanin, MW 3150 Da; and (C) bovine insulin, MW 5731 Da), 2-NPG matrix allows detection of higher charge states than 2,5-DHAP matrix. 2-NPG requires less laser power (ca. 4.2 J cm−2) than 2,5-DHAP (ca. 7.3 J cm−2) to obtain good ion abundance while maintaining higher charge states (Figure S4). However, the ion abundance near the respective threshold for ion formation is then slightly lower with 2-NPG relative to 2,5-DHAP (Figure S3). The laser power dependence is in agreement with a previous study that found a higher laser power induces lower charge states using 2,5-DHAP matrix.42
Figure 1.
LSIV-IMS-MS (1) total mass spectra and (2) two-dimensional plot of drift time vs. m/z of N-acetylated MBP peptide (MW ~1833 Da) with 2-NPG matrix acquired using (I) ‘LSI settings’39,42 at (A) low ‘4.2 J cm−2’ and (B) higher ‘7.3 J cm−2’ laser power with an inset of the isotopic distribution of +4 ion, and (II) commercial ‘MALDI settings’39 on a Waters SYNAPT G2 MALDI source. Insets in (2) show isotopic distributions and drift times of MBP ion.
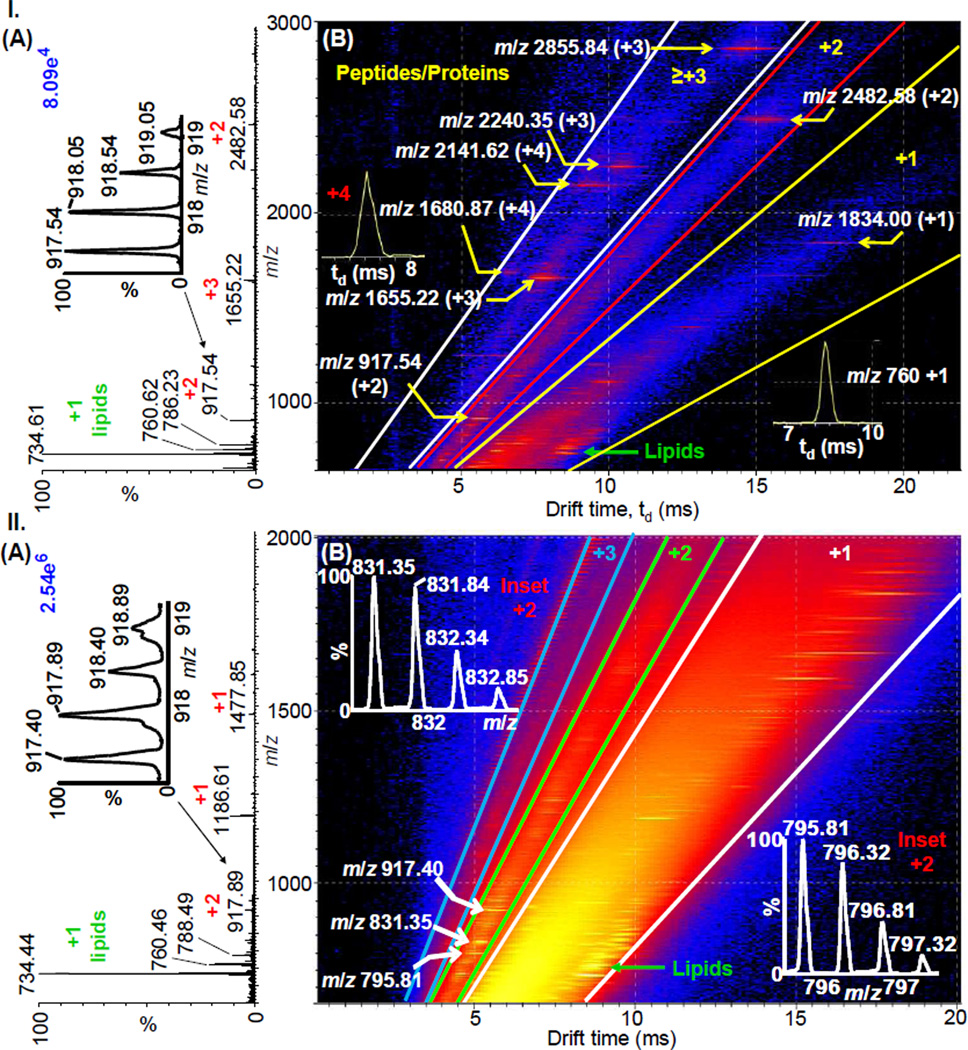
Figure 2 shows the LSIV analysis of delipified mouse brain tissue using ‘LSI settings’ with 2-NPG matrix applied to the tissue by different deposition methods. By directly spotting 100% 2-NPG matrix on the tissue and the use of a relatively high laser power (12.5 J cm−2), abundant multiply charged ions from peptides and proteins are obtained (Figure 2I.A) and are well separated by IMS from the singly charged lipid ions (Figure 2I.B). Ions are observed from proteins that are present in the tissue having molecular weights up to ~8.6 kDa (Figure S5) on a high performance mass spectrometer with a mass range of 8000. As noted above, spotting the tissue with 100% 2,5-DHAP matrix only detected up to ~3.2 kDa (Figure S1). Unfortunately, 100% 2-NPG matrix spray coated on a mouse brain tissue section required the application of a laser power of 12.5 J cm−2 to obtain sufficient ion abundance for good images (Figure 2II). Using ‘LSI settings’, as described above, under these higher laser fluence conditions, only +2 and +1 ions are detected (Figure 2II.A) from mouse brain tissue. However, the IMS dimension separating +2 from the +1 charge state allows extraction of the +2 peptide ions with good ion abundance (Figure 2II.B).
Figure 2.
LSIV-IMS-MS (A) total mass spectra and (B) two-dimensional plot of drift time vs. m/z of delipified mouse brain tissue (I) spotted and (II) spray coated with 100% 2-NPG matrix. Insets show (A) isotopic distributions of +2 charge state ion of the identified peptide MBP fragment, (I.B) drift times of charge state +4 and +1 ions, and (II.B) isotopic distribution of the +2 ions. Insets of higher charge states in (I) are included in Figure S5.
Just as in MALDI imaging, sample preparation is important with LSI imaging. Thus, a number of sample preparation protocols were explored. For example, mouse brain tissue sections mounted on a CHCA precoated glass slide, similar to previous protein studies,31 and spray coated with a binary mixture of 90% 2,5-DHAP and 10% 2-NPG allowed observation of higher molecular weight peptides and proteins ions (Figure S6) but with notably less ion abundance relative to 100% 2-NPG (Figure 2I). Thus, in the absence of the heated inlet27–40, the volatility of the matrix composition in vacuum defines the outcome of the tissue analysis and is especially restrictive to imaging for which direct matrix spotting cannot be employed because of delocalization of the tissue composition, as is also the case with MALDI. Clearly, further developments in sample preparation will be necessary to advance LSIV imaging just as it was for MALDI.
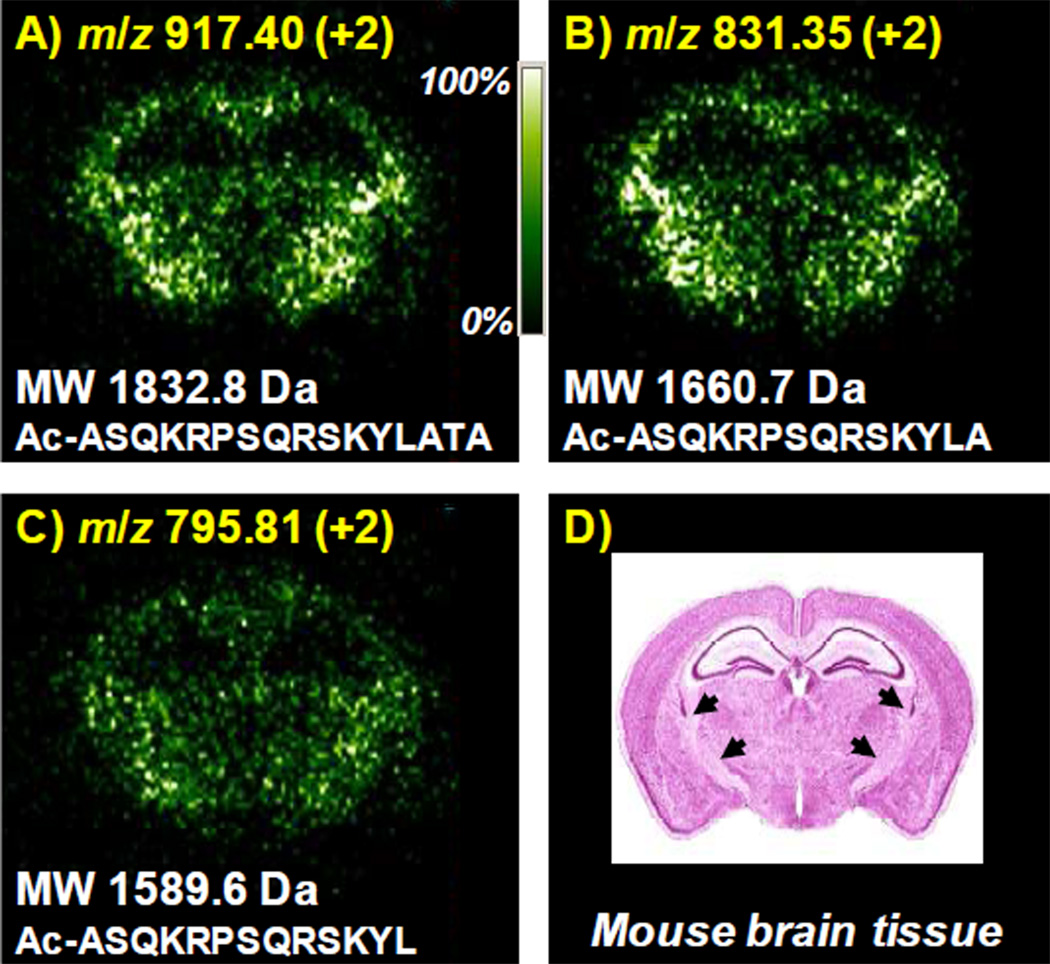
The drift time and m/z values of an area of choice can be selected in the 2-D IMS-MS plot, which becomes possible because of the two dimensionality of the dataset,52,53 allowing investigation of any ion of interest, while disregarding others. Here, using the ion mobility embedded mass spectral dataset from the 100% 2-NPG spray coating approach in Figure 2II.B, images were created for three peptides. This ion at m/z 917.40 had previously been sequenced using ETD fragmentation of the doubly charged N-acetylated MBP peptide (1–16) ion.31 The image (Figure 3A) relates well with the known location of the myelin basic protein (Figure 3D),54 validating our tissue imaging MS approach of multiply charged ions at vacuum. Using similar procedures, images of two additional doubly charged peptide ions are obtained, m/z 831.35 and 795.81 (Figure 3B,C), respectively. All three peptide images have the same location as the myelin basic protein in the mouse brain tissue (Figure 3D).54 Using the amino acid sequence information obtained from the identified peptide MBP fragment with a MW of 1832.8 Da31 (aa residues (1–16), Figure 3A), images of MW’s 1660.7 Da (aa residues (1–14), Figure 3B) and 1589.6 Da (aa residues (1–13), Figure 3C) then correspond to MBP peptides with two and three amino acid truncations (Figure 3B,C) of the C-terminal end.
Figure 3.
LSIV at intermediate pressure images of endogenous peptides from delipified mouse brain tissue spray coated with 100% 2-NPG matrix: A) m/z 917.40 (+2) MW 1832.8, B) m/z 831.35 (+2) MW 1660.7, and C) m/z 795.81 (+2) MW 1589.6, and (D) Coronal section of the mouse brain54 at a level similar to those analyzed in A-C. Arrowheads in (D) indicate heavily myelinated fiber tracts. Insets show molecular weight and amino acid sequence of the images relative to the identified peptide MBP fragment. The highest ion intensities are m/z 917 = 1.02 e4, m/z 831 = 1.27 e4, and m/z 795 = 1.06 e4, respectively.
Conclusions
Potential advantages of the LSI approach of tissue imaging include enhanced IMS gasphase separation of lipids, peptides, and proteins directly from mouse brain tissue. The extended mass range afforded by multiply charged ions makes it likely that imaging MS of multiply charged protein ions can be introduced with optimized sample preparation protocols and more appropriate desolvation conditions obtainable with physical instrument modifications. Similarly, it is expected that advanced vacuum fragmentation technology enabled by multiply charged ions, similar to LSI at atmospheric pressure,31 will improve sequence coverage for characterization of peptides and intact proteins, potentially even without the need for delipification by employing IMS gas-phase separation. The laser aligned in transmission geometry should enhance spatial resolution. Important strides toward more comprehensive tissue composition characterization relative to molecular location and structure determination using IMS-MS48–51 can be envisioned for imaging using multiply charged ions at vacuum.
Supplementary Material
Acknowledgements
Funding from Wayne State University (Schaap and Summer Dissertation Fellowships to EDI, and Schaap Faculty Scholar to ST), NSF CAREER 0955975, ASMS Research, DuPont Young Professor Award, and Eli Lilly Young Investigator Award in Analytical Chemistry (to ST) as well as DA011322 and DA021696 (to KM) are acknowledged. The authors are thankful to Waters Corporation for their support.
References
- 1.Karas M, Hillenkamp F. Anal. Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 2.Garrett TJ, Yost RA. Anal. Chem. 2006;78:2465–2469. doi: 10.1021/ac0522761. [DOI] [PubMed] [Google Scholar]
- 3.Laiko VV, Baldwin MA, Burlingame AL. Anal. Chem. 2000;72:652–657. doi: 10.1021/ac990998k. [DOI] [PubMed] [Google Scholar]
- 4.Senko SW, McLafferty FW. Annu. Rev. Biophys. Biomol. Struct. 1994;23:763–785. doi: 10.1146/annurev.bb.23.060194.003555. [DOI] [PubMed] [Google Scholar]
- 5.Vidova V, Po l J, Volny M, Novak P, Havlicek V, Wiedmer SK, Holopainen JM. J. Lipid Res. 2010;51:2295–2302. doi: 10.1194/jlr.M005488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ridenour WB, Kliman M, McLean JA, Caprioli RM. Anal. Chem. 2010;82:1881–1889. doi: 10.1021/ac9026115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tennessen JA, Woodhams DC, Chaurand P, Reiert LK, Billheimer D, Shyr Y, Caprioli RM, Blouin MS, Rollins-Smith LA. Dev. Comp. Immunol. 2009;33:1247–1257. doi: 10.1016/j.dci.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Remoortere A, van Zeijl RJM, van den Oever N, Franck J, Longuespee R, Wisztorski M, Salzet M. J. Am. Soc. Mass Spectrom. 2010;21:1922–1929. doi: 10.1016/j.jasms.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Trimpin S, Herath TN, Inutan ED, Wager-Miller J, Kowalski P, Claude E, Walker JM, Mackie K. Anal. Chem. 2010;82:359–367. doi: 10.1021/ac902065u. [DOI] [PubMed] [Google Scholar]
- 10.Rohner TC, Staab D, Stoeckli M. Mech. Ageing Dev. 2005;126:177–185. doi: 10.1016/j.mad.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 11.Pierson J, Norris Jl, Aerni HR, Svenningsson P, Caprioli RM, Andren PE. J. Proteome Res. 2004;3:289–295. doi: 10.1021/pr0499747. [DOI] [PubMed] [Google Scholar]
- 12.Stauber J, Lemaire R, Franck J, Bonnel D, Croix D, Day R, Wisztorski M, Fournier I, Salzet M. J. Proteome Res. 2008;7:969–978. doi: 10.1021/pr070464x. [DOI] [PubMed] [Google Scholar]
- 13.Chaurand P, Schwartz SA, Caprioli RM. J. Proteome Res. 2004;3:245–252. doi: 10.1021/pr0341282. [DOI] [PubMed] [Google Scholar]
- 14.Schwamborn K, Caprioli RM. Nature Reviews Cancer. 2010;10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- 15.Rauser S, Marquardt C, Balluff B, Deininger S, Albers C, Belau E, Hartmer R, Suckau D, Specht K, Ebert MP, Schmitt M, Aubele M, Hofler H, Walch A. J. Proteome Res. 2010;9:1854–1863. doi: 10.1021/pr901008d. [DOI] [PubMed] [Google Scholar]
- 16.Chandra S, Lorey DR. Cell Mol. Biol. 2001;47:503–518. [PubMed] [Google Scholar]
- 17.Chandra S, Lorey DR, II, Smith DR. Radiation Research. 2002;157:700–710. doi: 10.1667/0033-7587(2002)157[0700:qssims]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 18.Sjovall P, Lausmaa J, Johansson B. Anal. Chem. 2004;76:4271–4278. doi: 10.1021/ac049389p. [DOI] [PubMed] [Google Scholar]
- 19.Touboul D, Kollmer F, Niehuis E, Brunelle A, Laprevote O. J. Am. Soc. Mass Spectrom. 2005;16:1608–1618. doi: 10.1016/j.jasms.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Jones EA, Lockyer NP, Vickerman JC. Int. J. Mass Spectrom. 2007;260:146–157. [Google Scholar]
- 21.Maarten Altelaar AF, van Minnen J, Jiméez CR, Heeren RMA, Piersma SR. Anal. Chem. 2005;77:735–741. doi: 10.1021/ac048329g. [DOI] [PubMed] [Google Scholar]
- 22.McDonnel L, Piersma SR, Maarten Altelaar AF, Mize TH, Luxembourg SL, Verhaert PDEM, van Minnen J, Heeren RMA. J. Mass Spectrom. 2005;40:160–168. doi: 10.1002/jms.735. [DOI] [PubMed] [Google Scholar]
- 23.Altelaar AFM, Klinkert I, Jalink K, de Lange RPJ, Adan RAH, Heeren RMA, Piersma SR. Anal. Chem. 2006;78:734–742. doi: 10.1021/ac0513111. [DOI] [PubMed] [Google Scholar]
- 24.Wiseman JM, Puolitaival SM, Takats Z, Cooks RG, Caprioli RM. Angew. Chem. Int. Ed. 2005;44:7094–7097. doi: 10.1002/anie.200502362. [DOI] [PubMed] [Google Scholar]
- 25.Wiseman JM, Ifa DR, Song Q, Cooks RG. Angew. Chem. Int. Ed. 2006;45:7188–7192. doi: 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]
- 26.Wiseman JM, Ifa DR, Zhuc Y, Kissingerc CB, Manicke NE, Kissingera PT, Cooks RG. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18120–18125. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trimpin S, Herath TN, Inutan ED, Cernat SA, Wager-Miller J, Mackie K, Walker JM. Rapid Commun. Mass Spectrom. 2009;23:3023–3027. doi: 10.1002/rcm.4213. [DOI] [PubMed] [Google Scholar]
- 28.Trimpin S, Inutan ED, Herath TN, McEwen CN. Anal. Chem. 2010;82:11–15. doi: 10.1021/ac902066s. [DOI] [PubMed] [Google Scholar]
- 29.Trimpin S, Inutan ED, Herath TN, McEwen CN. Mol. Cell. Proteomics. 2010;9:362–367. doi: 10.1074/mcp.M900527-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards AL, Marshall DD, Inutan ED, McEwen CN, Trimpin S. Rapid Commun. Mass Spectrom. 2011;25:1–4. doi: 10.1002/rcm.4841. [DOI] [PubMed] [Google Scholar]
- 31.Inutan ED, Richards AL, Wager-Miller J, Mackie K, McEwen CN, Trimpin S. Mol. Cell. Proteomics. 2011;10:1–8. doi: 10.1074/mcp.M110.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inutan ED, Trimpin S. J. Am. Soc. Mass Spectrom. 2010;21:1260–1264. doi: 10.1016/j.jasms.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 33.Inutan ED, Trimpin S. J. Proteome Res. 2010;9:6077–6081. doi: 10.1021/pr1005923. [DOI] [PubMed] [Google Scholar]
- 34.Richards AL, Lietz CB, Wager-Miller J, Mackie K, Trimpin S. Rapid Commun. Mass Spectrom. 2011;25:815–820. doi: 10.1002/rcm.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards AL, Lietz CB, Wager-Miller J, Mackie K, Trimpin S. J. Lipid Res. 2012 doi: 10.1194/jlr.D019711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McEwen CN, Pagnotti V, Inutan ED, Trimpin S. Anal. Chem. 2010;82:9164–9168. doi: 10.1021/ac102339y. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Inutan ED, Wang B, Richards AL, Lietz CB, Green DR, Manly CD, Richards AL, Marshall DD, Lingenfelter S, Ren Y, Trimpin S. J. Am. Soc. Mass Spectrom. 2012 doi: 10.1007/s13361-012-0413-z. [DOI] [PubMed] [Google Scholar]
- 38.McEwen CN, Trimpin S. Int. J. Mass Spectrom. 2011;300:167–172. [Google Scholar]
- 39.Trimpin S, Wang B, Inutan ED, Li J, Lietz CB, Pagnotti VS, Harron AF, Sardelis D, McEwen CN. J. Am. Soc. Mass Spectrom. 2012 doi: 10.1007/s13361-012-0414-y. [DOI] [PubMed] [Google Scholar]
- 40.Trimpin S, Ren Y, Wang B, Lietz CB, Richards AL, Marshall DD, Inutan ED. Anal. Chem. 2011;83:5469–5475. doi: 10.1021/ac2007976. [DOI] [PubMed] [Google Scholar]
- 41.McEwen CN, Larsen BS, Trimpin S. Anal. Chem. 2010;82:4998–5001. doi: 10.1021/ac1006624. [DOI] [PubMed] [Google Scholar]
- 42.Inutan ED, Wang B, Trimpin S. Anal. Chem. 2011;83:4076–4084. doi: 10.1021/ac2000576. [DOI] [PubMed] [Google Scholar]
- 43.Lietz CB, Richards AL, Ren Y, Trimpin S. Rapid Commun. Mass Spectrom. 2011;25:3453–3456. doi: 10.1002/rcm.5233. [DOI] [PubMed] [Google Scholar]
- 44.Iribarne JB, Thomson BA. J. Chem. Phys. 1976;64:2287–2289. [Google Scholar]
- 45.Dole M, Mack LL, Hines RL, Mobley RC, Ferguson LD, Alice MB. J. Chem. Phys. 1968;49:2240–2249. [Google Scholar]
- 46.Frankevich V, Nieckarz R, Sagulenko P, Barylyuk K, Levitsky L, Agapov A, Perlova T, Gorshkov M, Tarasova I, Zenobi R. Rapid Commun. Mass Spectrom. 2012;26:1567–1572. doi: 10.1002/rcm.6264. [DOI] [PubMed] [Google Scholar]
- 47.Zhigilei LV, Garrison BJ. J. Appl. Phys. 2000;88:1281–1298. [Google Scholar]
- 48.Clemmer DE, Jarrold MF. J. Mass Spectrom. 1997;32:577–592. [Google Scholar]
- 49.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. Int. J. Mass Spectrom. 2007;261:1–12. [Google Scholar]
- 50.McLean JA, Ruotolo BT, Gillig KJ, Russell DH. Int. J. Mass Spectrom. 2005;240:301–315. [Google Scholar]
- 51.Wyttenbach T, Bowers MT. Top. Curr. Chem. 2003;225:207–232. [Google Scholar]
- 52.Trimpin S, Clemmer DE. Anal. Chem. 2008;80:9073–9083. doi: 10.1021/ac801573n. [DOI] [PubMed] [Google Scholar]
- 53.Trimpin S, Tan B, Bohrer BC, O'Dell DK, Merenbloom SI, Pazos MX, Clemmer DE, Walker JM. Int. J. Mass Spectrom. 2009;287:58–69. [Google Scholar]
- 54.Sidman RL, Kosaras B, Misra BM, Senft SL. [Accessed [June 9, 2012]];High Resolution Mouse Brain Atlas. 1999 at http://www.hms.harvard.edu/research/brain/atlas.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.