Figure 1.
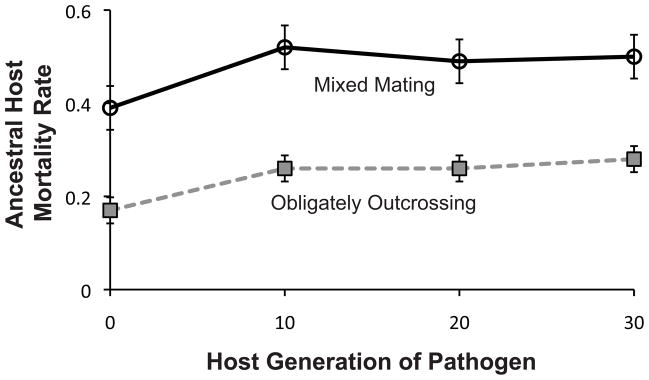
Pathogen infectivity over time. S. marcescens populations coevolved with either mixed mating or obligately outcrossing C. elegans were sampled every ten generations of coevolution. These pathogens were used to infect ancestral populations of the hosts with which they were coevolving, thus measuring their infectivity over time. The pathogen populations show an initial increase and then maintenance of infectivity throughout the experiment, as evidenced by the increase (generation 0 to generation 10 ; mixed mating, F1,12= 13.69, p = 0.003; obligately outcrossing, F1,12 = 79.79, p < 0.0001) and maintenance (generation 10 to generation 30; mixed mating, F1,12 = 0.4547, p = 0.5129; obligately outcrossing, F1,12 = 0.6826, p = 0.4248) of host mortality rates against the same ancestral hosts. Error bars indicate ± 1 S.E.M.
