Fig. 5.
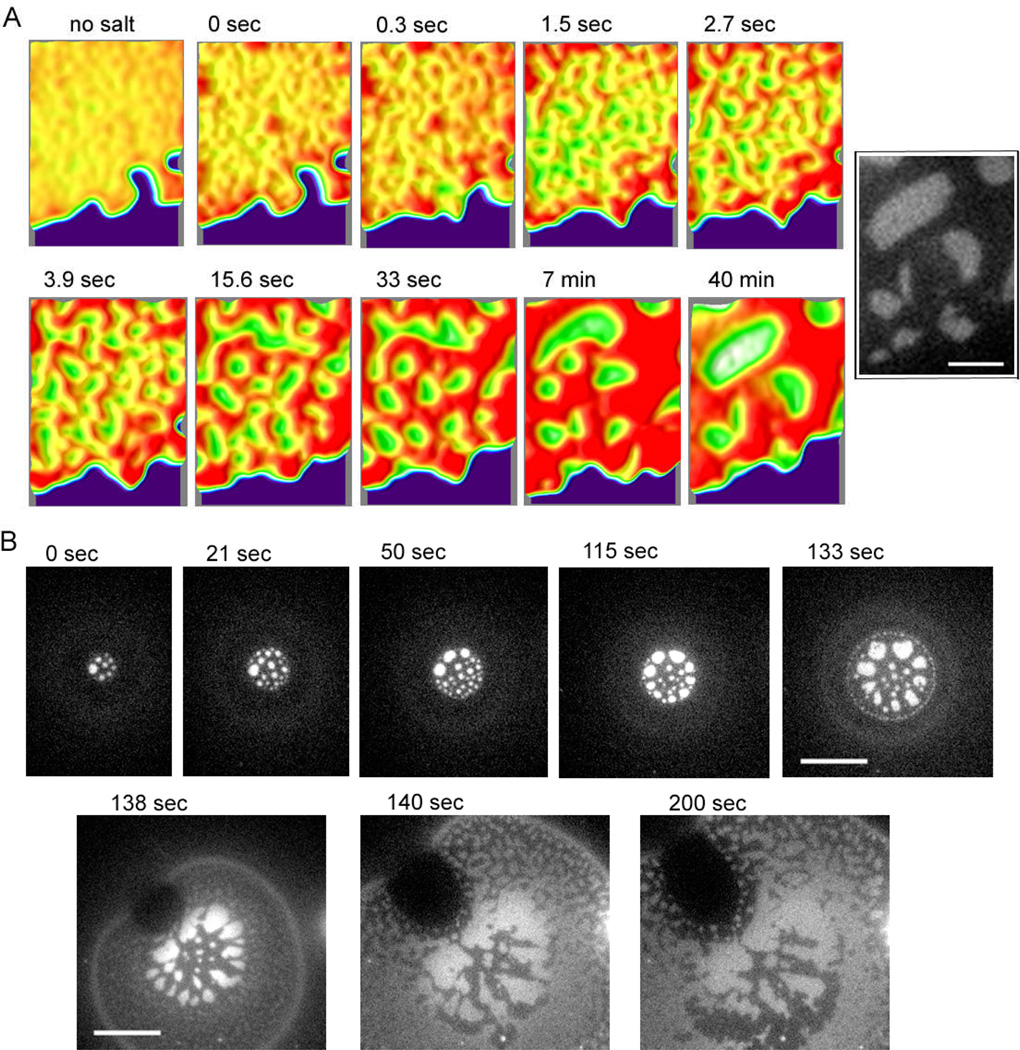
Topological pattern evolution process during GUV adhesion and patch formation monitored with Alexa 488 labelled 24mer DNA by epi-fluorescence microscopy. GUVs displaying Alexa 488 labelled 24mer and unlabelled 48mer were bound on a supported bilayer displaying the complementary DNA where they flatten and rupture to form patches (see Fig. 1). (A) A tethered patch was formed, then the DNA tethers were un-hybridized by removing salt, and DNA hybridization was re-initiated by adding salt so that initial state of segregation in the tethered patches could be observed under controlled conditions. The DNAs become uniformly distributed after several minutes incubation in deionized water (no salt), then the salt was added, and after a few seconds, the patch became slightly rough, which is set as 0 sec. The segregation by tether length occurred rapidly, forming small 24mer (bright) and 48mer domains (~ 3 sec). The randomly distributed small domains gradually merged and became rounder in shape (~ 7 min); this segregation process continued, but became very slow after 10 min. The surface topology of the segregated patch is reconstituted for enhanced contrast by using ImageJ (v2.41) interactive 3D surface plot plug-in with thermal colour scale (red is high and blue is low). Inverted brightness was used for the patch area to depict realistic domain height – making the higher 48mer region brighter. The black and white inset is the original image at 40 min for reference. (B) Initial domain formation process as a GUV flattens and the binding area expands. Initially, circular bright 24mer domains were generated, arranged roughly concentrically (0~133sec). After the GUV ruptured and formed a patch (see Fig. 1), the circular 24mer domains became amorphous, and small segregated topological domains develop in new binding regions (138 sec). The new domains then started to merge with the already formed domains inside (~200 sec). Scale bar is 3 µm for (A) and 10 µm for (B).