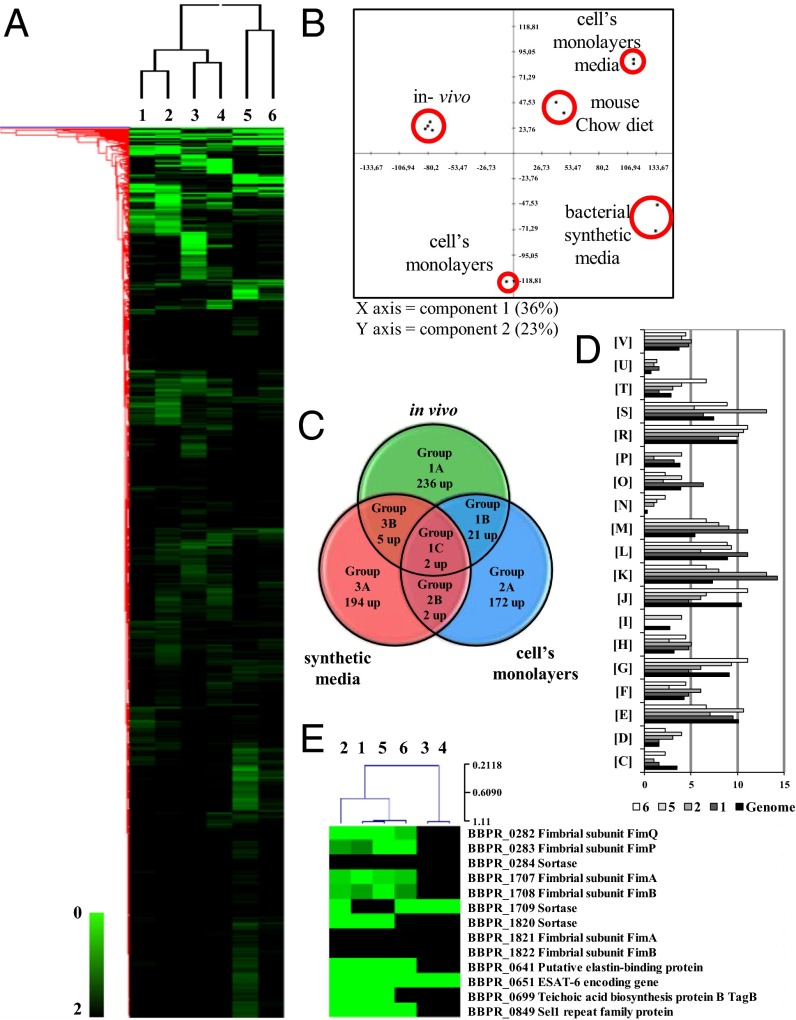
Fig. 1.
Identification of B. bifidum PRL2010 differentially expressed genes by transcriptome analysis in response to contact with the host. (A) Heat-map displaying the change in PRL2010 gene expression upon colonization of murine caeca (lanes 1 and 2), when grown in DMEM synthetic medium (lanes 3) or in fresh chow diet (lane 4), and following incubation with human intestinal HT29 cells (lanes 5 and 6). Each row represents a separate transcript and each column represents a separate sample. Color legend is on the bottom of the microarray plot; green indicates increased transcription levels compared with the reference samples. The reference conditions used were as follows: lane 1, fresh chow diet; lanes 2–4 and 6, MRS medium; lane 5, DMEM. Dendrogram on the left margin of the heat-map represents the hierarchical clustering algorithm result based on average linkage and Euclidean distance of the gene dataset. (B) The clustering of PRL2010 transcriptomes under in vitro and in vivo conditions by principal component analysis. (C) Venn diagram showing the number of genes expressed during the different conditions: in vitro (exposure to a human cell line), in vivo, and DMEM synthetic medium. (D) Depiction of a functional annotation of the in vitro- and in vivo-expressed genes of B. bifidum PRL2010 according to their COG categories. Each COG family is identified by a one-letter abbreviation (National Center for Biotechnology Information database). For each category, the black bar represents the percentage of genes in that category as detected in the sequenced genome of PRL2010 (7). The other bar shows the percentages of genes transcribed during murine colonization by PRL2010 (conditions 1 and 2), and following exposure of PRL2010 cells to human intestinal cells (conditions 5 and 6). The percentage was calculated as the percentage of transcribed genes belonging to the indicated COG category with respect to all transcribed genes. (E) Selected genes that were up-regulated when PRL2010 were cultivated in the conditions indicated in A.