Abstract
Intravascular optical coherence tomography (IVOCT) is rapidly becoming the method of choice for the in vivo investigation of coronary artery disease. While IVOCT visualizes atherosclerotic plaques with a resolution <20µm, image analysis in terms of tissue composition is currently performed by a time-consuming manual procedure based on the qualitative interpretation of image features. We illustrate an algorithm for the automated and systematic characterization of IVOCT atherosclerotic tissue. The proposed method consists in a supervised classification of image pixels according to textural features combined with the estimated value of the optical attenuation coefficient. IVOCT images of 64 plaques, from 49 in vivo IVOCT data sets, constituted the algorithm’s training and testing data sets. Validation was obtained by comparing automated analysis results to the manual assessment of atherosclerotic plaques. An overall pixel-wise accuracy of 81.5% with a classification feasibility of 76.5% and per-class accuracy of 89.5%, 72.1% and 79.5% for fibrotic, calcified and lipid-rich tissue respectively, was found. Moreover, measured optical properties were in agreement with previous results reported in literature. As such, an algorithm for automated tissue characterization was developed and validated using in vivo human data, suggesting that it can be applied to clinical IVOCT data. This might be an important step towards the integration of IVOCT in cardiovascular research and routine clinical practice.
OCIS codes: (100.0100) Image processing; (100.2960) Image analysis; (100.4995) Pattern recognition, metrics; (170.0170) Medical optics and biotechnology; (170.6935) Tissue characterization
1. Introduction
Coronary atherosclerotic plaques are mainly divided into lipid-rich, calcified and fibrous types, with mixed forms appearing in many cases. Plaque composition and morphology is strongly correlated with the clinical presentation of the patient. Calcified plaques (CP) predominantly occur in stable lesions while acute coronary syndromes are typically caused by the rupture of a so called ‘vulnerable’ plaque, the TCFA (thin-cap fibroatheroma), consisting of an accumulation of extracellular lipid with a thin overlying fibrous cap [1,2]. Moreover, specific types of coronary plaques have been shown to impact the outcome of percutaneous coronary intervention (PCI) procedures [3–6].
Detailed identification and characterization of atherosclerotic lesions in vivo and quantification of relative morphological, chemical and mechanical parameters contributes in advancing the general understanding of coronary atherosclerotic disease and facilitating the development of new therapies and interventions. However, most of the current available invivo imaging modalities, such as intravascular ultrasound (IVUS) and computed tomography (CT), are limited by a low spatial resolution (>100µm) [7,8].
Intravascular optical coherence tomography (IVOCT) is a catheter based imaging modality able to visualize the arterial wall from the luminal side with an axial resolution <20µm and a penetration depth of ~0.5-1.5 mm [9]. State-of-the art systems are able to acquire IVOCT images at a frame rate of 100 frames per second imaging a complete arterial volume with a length of ~5.4 cm in only a few seconds.
Previous studies demonstrated the ability of IVOCT to qualitatively characterize the three major plaque components by correlation with histology [10,11]. Yabushita et al. [11] described fibrous plaques as homogeneous signal-rich regions; calcified plaques as heterogeneous signal-poor regions with sharply delineated borders and lipid-rich plaques as signal-poor regions with diffuse borders (Fig. 1). A more recent publication by Xu et al. [12] quantified the true backscattering coefficient µb and the attenuation coefficient µt of atherosclerotic tissues (Table 1), corroborating the qualitative scheme for IVOCT plaques assessment proposed by the former study [11]. However, the manual assessment of IVOCT plaque tissue components based on qualitative criteria leads to interobserver variability [13]. Furthermore, manual tracing and assessment of plaques in IVOCT images is a very time-consuming and labor intensive procedure. As such, methods for automated IVOCT tissue characterization would be a very important step for a widespread integration of this technology in cardiovascular research and clinical practice.
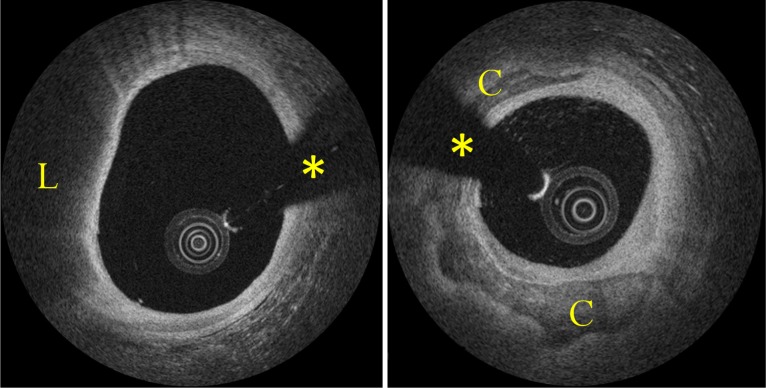
Fig. 1.
Examples of atherosclerotic plaques. Left image shows a lipid-rich plaque (L); right image a circumferential calcification (C). Asterisk indicates guide-wire shadowing artifact.
Table 1. Attenuation and Backscattering for the Main Atherosclerotic Plaque Components [12].
| Plaque type | Backscattering | Attenuation |
|---|---|---|
| Fibrous | High | Low |
| Lipid | High+ | High |
| Calcium | Low | Low- |
To overcome some of these problems, van Soest et al. [14] proposed an algorithm for the automatic quantification of the optical attenuation coefficient in IVOCT images. Such a method allows for discrimination of high attenuating plaque components (i.e. lipid core and macrophages infiltrations) from low attenuating components (i.e. fibrous and calcified tissue). Another recent publication [15] presented an algorithm for the semiautomatic segmentation of calcified plaques.
In this study, we propose an algorithm for the automated characterization of the three main plaque components: fibrotic, calcified and lipid-rich tissue. Qualitative features of atherosclerotic plaques described by former studies [11,16] are quantified through statistical texture analysis as well as information related to tissue backscattering [17]. In addition, as the attenuation coefficient µt is a strong discriminant between lipid and other atherosclerotic tissues [12,14], the local tissue attenuation is quantified by means of a novel algorithm. All the extracted features are then combined in a supervised classification algorithm resulting in an automated characterization of atherosclerotic plaques. The classification algorithm istrained by a set of data visualizing atherosclerotic tissue from 24 different plaques visualized in vivo. The performance of the algorithm is subsequently evaluated against the manual assessment of a testing set composed by 40 in vivo atherosclerotic plaques by an expert IVOCT reader. Measured optical properties are compared to previous results reported in literature.
2. OCT system
Intracoronary images employed in this study were collected with a state-of-the-art commercially available Fourier-Domain OCT system (C7-XR, St. Jude Medical, St. Paul, Minnesota) by means of the C7 Dragonfly intravascular OCT catheter (St. Jude). The system is equipped with a near-infrared laser light source with a central wavelength of 1305 nm and a scan range of ± 80 nm. The sample arm contains an objective lens with a low numerical aperture (NA) of approximately 0.3. Imaging specifications, according to the manufacturer, consist in an axial resolution <20 µm in biological tissues and a lateral resolution of 25-60 µm. Scan parameters are 100 frames per second, 54,000 A-scans per second, pullback speed of 20 mm/sec, pullback length of ~54 mm and scan-range (in contrast medium) of ~4.8 mm.
3. Classification algorithm
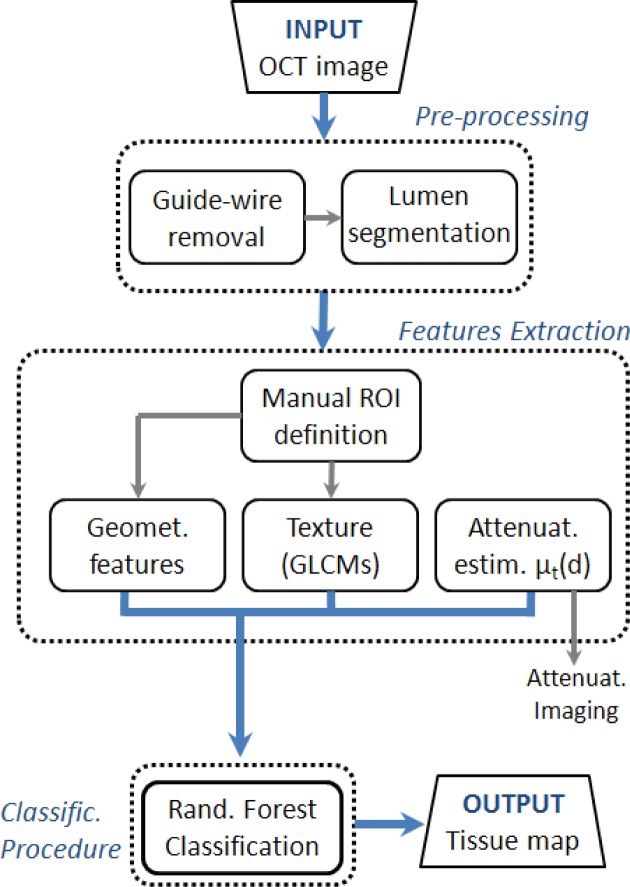
Figure 2 presents a flow chart of the entire method for atherosclerotic tissue characterization. After a first preprocessing step, including automatic guide-wire and lumen segmentation, the local attenuation coefficient µt(d) is estimated and local gray scale features and geometrical features quantified. Based on these locally extracted values, supervised pixel classification is applied. The output of the algorithm provides a color-coded image map depicting the different types of tissue. Image processing is applied to IVOCT data in the polar image domain.
Fig. 2.
Flowchart of the image processing algorithm for tissue characterization. Output consists of a tissue image map. Intermediate output is given by the attenuation image-map.
3.1 Preprocessing
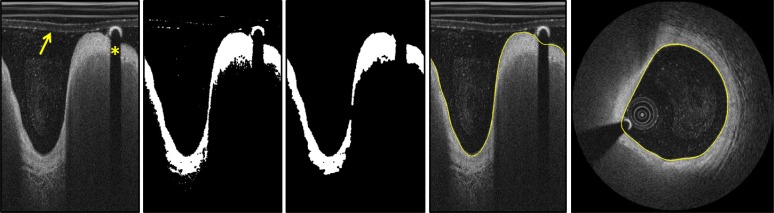
Guide-wire and catheter artifacts are automatically removed by applying an area constraint: the polar IVOCT image is converted to a binary image using the Otsu’s method [18] and made uniform through a morphological closing operation [15,19]. Subsequently, the guide-wire and the imaging catheter are recognized as unconnected regions with a pixel areasmaller than a predefined threshold BWMA (black-white minimal area) (Fig. 3) and the lumen is automatically segmented using an algorithm previously developed and validated by our research group [20].
Fig. 3.
Illustration of the fully-automated lumen segmentation procedure. Image (a) shows the polar domain image. Image (b) illustrates the application of the Otsu’s method and morphological operations and image (c) results of the area constrain. Image (d) and (e) show segmentation results before and after scan-conversion respectively. On image (a) the arrow is pointing to the OCT imaging catheter and the asterisk indicates the guide-wire shadowing artifact.
3.2 Feature extraction
3.2.1 Attenuation analysis
IVOCT systems acquire multiple OCT A-lines while retracting and rotating the catheter visualizing an entire portion of a vessel volume. Under the assumption that OCT only detects light that has been scattered once [21,22] and that the tissue is homogeneous, the intensity of such an A-line I(d) decreases with depth according to the Lambert-Beer law [14,23,24]:
| (1) |
where d indicates the penetration depth, µt the total attenuation coefficient (comprehending both absorption µa and scattering µs) and I0 a scale factor equal to the locally available intensity I’ multiplied by the backscattering coefficient µb [14]. In addition, the OCT signal is influenced by focusing effects related to the confocal properties of the OCT catheter [24–26], that can be described as:
| (2) |
where T(d) is the axial point spread function (PSF), x0 indicates the catheter focal point and z0 the Rayleigh range. As such, the OCT signal can be modeled as:
| (3) |
As biological tissues in general, the vessel wall is a heterogeneous structure. The healthy vessel wall is composed of different tissues (i.e. intima, media, adventitia and perivascular tissue). Similarly, atherosclerotic tissue is composed by multiple layers (e.g. fibrotic cap and calcified/lipidic tissue beneath). As such, an IVOCT A-line typically samples at least two or more tissue types. Previous studies from van der Meer et al. demonstrated that optical parameters can be fitted to different tissue layers through individual fitting [24,27]. This supports the fact that the diseased vessel wall can be considered a multi-layered structure and that Eq. (3) can be fitted through the different tissue layers. To automatically fit the proposed model to the different layers, we propose to model the signal I(d) as:
| (4) |
where i is the layer number and П(d–di) a rectangular window in depth of variable length:
| (5) |
where di is the i-layer starting position, t a constant value taking into account the transition between different layers and I0,i defined as:
| (6) |
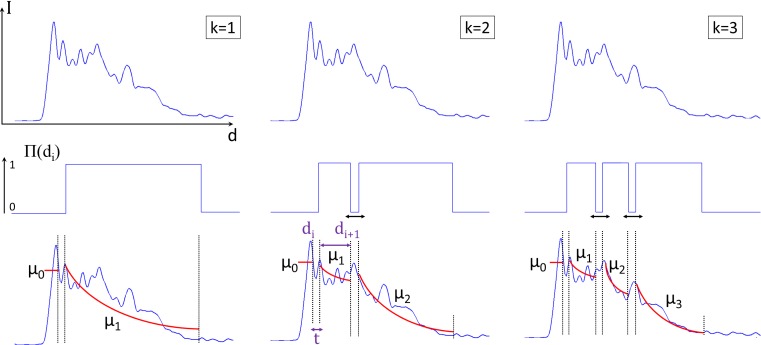
This signal model is iteratively fitted to the OCT A-lines for different values of k (i.e. number of layers) and for different positions (di) of the individual layers solving an optimization problem. This procedure is illustrated in Fig. 4. At every step k, all the possible fits are computed varying П(d-di) and the optimal solution at a step k, is computed as the fit presenting the minimum sum-of-square error (SSE) difference in respect to the OCT A-line. At the end of the procedure, the number of layers k best describing the signal is selected through the use of a threshold SSEmin: a higher number of layers k is considered to better fit the signal when the relative improvement over the fit k-1 is larger than SSEmin. Please note that this model thus not take the roll-off of the OCT system into account.
Fig. 4.
A schematic representation of the iterative fitting procedure. From this image it is possible to appreciate how the algorithm tests all the possible solutions for every value of k varying П(di).
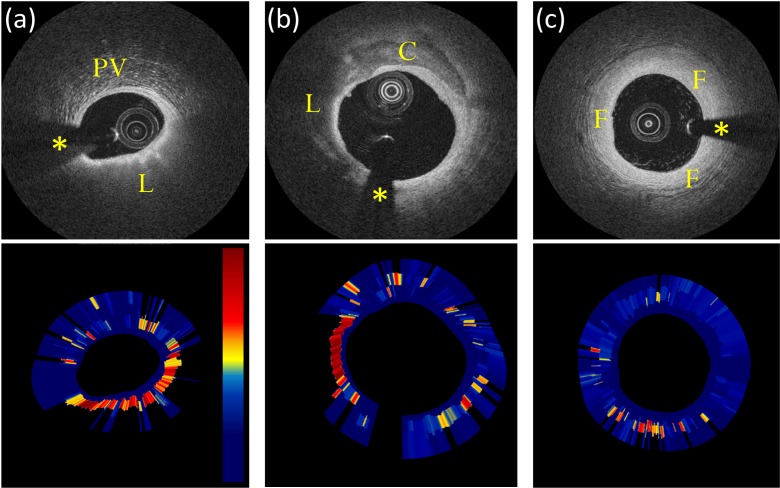
This attenuation analysis results in an automatic estimation of µt(d) for every pixel along each A-line (Fig. 5). The aim of this method is a robust identification of homogeneous portions of the signal, correctly dealing with the presence of sudden changes in intensity at different tissue interfaces as a result of differences in the backscattering coefficient µb.
Fig. 5.
Attenuation imaging examples. Image (a) contains a lipid-rich plaque (L) and peri-vascular tissue (PV). Image (b) contains a mixed atherosclerotic plaque containing both lipid-rich (L) and calcific (C) tissue; image (c) shows an example of intimal thickening (fibrotic tissue). Automatic analysis correctly depicts lipid-rich tissue with a higher attenuation coefficient. Attenuation images µt(d) are displayed on a color scale from 0 (dark blue) to 12 mm−1 (dark red). Asterisks indicate the guide-wire shadowing artifact.
3.2.2 Texture analysis
Spatial gray-level dependence matrices (SGLDMs), or co-occurrence matrices, are a well-known method for texture analysis [28,29]. SGLDMs represent the conditional joint probabilities of all pair-wise combinations of gray levels given two parameters: inter pixel distance (d) and orientation (θ). According to the literature [28,30], it is possible to extract 22 statistical features from SGLDMs for the description of textural properties of a kernel K of the analyzed image quantized on G gray-levels. In this study, we identified a reduced set of 7 features that can best distinguish between the different types of tissue components based on an analysis detailed in the Appendix. In addition, mean pixel intensity (first order statistic) was also quantified within the same kernel K and used as a classification feature as well as the distance of pixels of interest to the imaging catheter accounting for distance effect in the image features (e.g. due to system roll-off). Mathematical definitions of all the statistical features considered in this study are reported in Table 2.
Table 2. Definitions for the First and Second Order Statistical Features Used in This Study.
| Mean (1st order stat.) | where N is the number of pixels in a given region and Ik the int. value of a pixel | |
| Co-occurrence probability (2nd order stat.) | where Pi,j represents the number of occurrences of gray levels i and j for a certain (d, θ) pair in a given region | |
| Contrast | ||
| Cluster shade | ||
| Energy | ||
| Entropy* | ||
| Homogeneity | ||
| Information measures of correlation* | hij = entropy | |
| and | ||
| Inverse difference moment |
*eps is an arbitrarily small constant chosen to be 2−52 (floating point accuracy).
3.3 Pixel classification
All the previously extracted features are combined in a feature vector (FV). FV is a vector of dimensions n*m, with n (number of rows) equal to the number of pixels included in the analysis and m (number of columns) the number of features describing each pixel. This vector is used as input to a recently proposed supervised classification algorithm: Random Forests (RF) [31]. RF is an ensemble classifier that tries to achieve an accurate classification by combining a large number of weak classifiers (i.e. binary trees) thus presenting reduced computational costs. The main characteristic of RF is that the correlation between individual base learners is reduced by choosing the best predictors in a random subspace of the training data. RF combines multiple binary tree predictors where each tree is constituted by a randomly sampled subset of features of the training-set: each object to classify is processed by all the trees, every tree votes for a single class and the forest chooses the classification having the larger number of votes. Required parameters are ntree (number of trees to grow) and mrand (number of variables randomly sampled at each node). More specific details about the RF algorithm can be found on the original report [31] and subsequent publications [32–34].
Every row of FV is classified into four different classes: (1) fibrotic, (2) calcified, (3) lipidic tissue, (4) outliers and corresponding image pixels are subsequently color-coded. Hence, classification results can be displayed side-by-side to IVOCT standard images, resulting in a tissue color-map.
3.4 Implementation details
Algorithms for lumen segmentation, attenuation analysis and texture extraction were implemented in Matlab 7.12.0 (R2011a) (MathWorks, Natick, MA). The scan-conversion algorithm was implemented in C++ (Microsoft, Redmond, WA) and the Random Forests classification algorithm is based on the original FORTRAN (IBM, Armonk, NY) code made available by the RF inventors [31]. Both C++ and FORTRAN codes were executed in Matlab by the means of MEX libraries.
3.5 Attenuation algorithm implementation
The polar OCT image is first pre-processed with a 2D low-pass Gaussian filter of dimension hG and standard deviation σG for speckle reduction. Then the OCT signal is compensated for the effect of the axial PSF T(d) and subsequently linearized through loge-compression. This allows for a linear least-square fitting of the model with the advantage, over non-linear iterative algorithms, of independence from initialization [14]. Computational time reduction is another advantage. The fitting starting point ds is automatically located as the deepest pixel with intensity <95% of the maximum A-line intensity starting from the lumen-vessel interface (lumen segmentation) dL. The end of the A-line dend is defined according to the maximum penetration depth of IVOCT dmax [9,13]. During the fitting procedure, results presenting ‘positive’ attenuation values µt>0 are automatically set to a minimum attenuation value µt,min = 0 as this would be a non-physical solution. Moreover, any pixel behind an area of high attenuation reaching a predefined minimum signal-to-noise-ratio (SNR) is automatically excluded for further analysis.
3.6 Parameter tuning
Besides the training of the RF algorithm, training set images were used for optimal tuning of the algorithm parameters. The lumen segmentation parameter BWMA was empirically set to a fixed value of 1,500 pixels, which was kept constant for all processed images. Attenuation algorithm parameters were respectively set to: wmin = 255µm, tt = 25.5 µm (approximately 2 times the axial resolution of the system), the step size di was equal to 1 pixel, k = 4 and SSEmin = 2.5 (i.e. the minimal sum-of-square error difference between the fitted model and the OCT trace). SSEmin was also determined empirically, by testing a wide range of different values over a large number of different A-scan lines. The value of k represents the maximum number of layers to be fitted. Minimum SNR was determined as the mean intensity value of image regions with no tissue or other objects present. According to previous studies [9,13], dmax was set to be equal to 1.5 mm. Attenuation preprocessing parameters hG and σG were set equal to 7 pixels and 2.5 respectively. Nominal Dragonfly catheter parameters (St. Jude), for longitudinal PSF signal compensation, were characterized to be: x0≈1.5 mm (focal point) and z0≈2 mm (Rayleigh length). Statistical features analysis parameters were set to d = 2 pixels, θ = 0° and 90°, K = 11 pixels and G = 64 as detailed in the Appendix. Random Forests algorithm parameters were set to ntree = 120 and mrand = 10.
4. Data collection
4.1 IVOCT images
The IVOCT data utilized in this study were acquired in the context of two clinical trials, STACCATO (NCT01065519) and SEDUCE (NCT01065532) (University Hospitals Leuven, Belgium). These studies comply with the declaration of Helsinki and were approved by the local ethic committee. All patients gave informed consent.
Overall, IVOCT images of evident atherosclerotic plaques from 49 different pullbacks from 49 different patients were arbitrarily selected from our database as follows: data sets were read by an expert and 64 different plaques were visually located. Then, for every selected plaque, only one single representative IVOCT image was retained in order to avoid redundant (i.e. correlated) information in the training- and testing- sets. This resulted in a total of 64 images of 64 different atherosclerotic plaques, generating an appropriate set of images for algorithm training and validation.
Polar raw-image data exported from the C7 system were used for automatic image analysis. Images were scan-converted with an in-house developed algorithm [20,35] to standard IVOCT Cartesian domain for the purpose of manual analysis.
4.2 Manual tissue classification
All images were analyzed by an expert IVOCT reader and atherosclerotic plaques were manually identified and classified using the software ImageJ [36,37] according to the qualitative criteria previously validated [11,12]. The normal artery wall –characterized by a very thin three-layered intima, media and adventitia architecture [16]– as well as the surrounding peri-vascular tissue were excluded from the analysis by defining a region of interest (ROI) within the IVOCT maximum penetration depth. Based on manual classification, pixels were labeled as: fibrous, calcified or lipid-rich. In addition, guide-wire shadowing, catheter, vessel lumen, unknown tissue and pixels exceeding the maximum IVOCT penetration depth were labeled as ‘outliers’. An average number >500 pixels per image was retained. As such, a balanced training set composed of 3,100 pixels per class (4 in total), corresponding to 12,400 pixels overall, was created and stored.
5. Training and validation
24 images from the 64 images mentioned above were selected to create the training set. This training set was used both to tune algorithm parameters (cf. section 3.6) and to train the classifier. The remaining 40 images were used as test set and the performance of the automated classification algorithm was evaluated on these images. In addition, adjacent frames of a randomly selected plaque were analyzed to test classification consistency.
Classification accuracy was computed within the ROIs manually identified by the expert OCT reader to avoid bias, by comparing the ground-truth pixels (manual analysis) to the automated algorithm results. Accuracy was computed as the percentage of image pixels assigned to the correct class label [38,39]. Both per-class and global classification accuracy were quantified. Moreover, the percentage of pixels automatically excluded from analysis by the algorithm with respect to the ground truth, was quantified and reported as ‘classification feasibility’. In addition, measured values of the classification parameters were compared to literature when possible. In addition, statistical significance of parameters was tested through one-way ANOVA for multiple comparisons (and Bonferroni post-hoc testing when appropriate) and their relative importance for classification was quantified [31].
6. Results
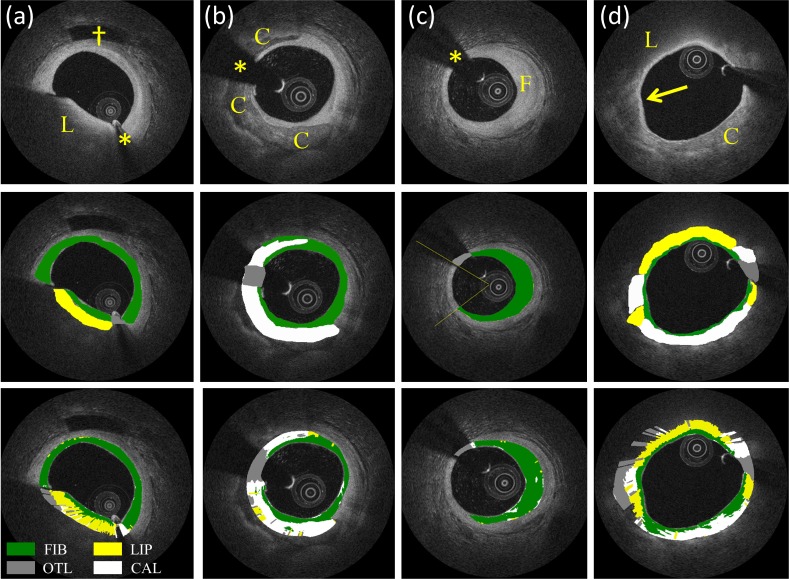
Figure 6 shows examples of the automated classification results (lowest panels) for lipid, calcified, fibrotic and mixed plaques, respectively, as well as manual classification (middle panels). Figure 7 shows the results of the classification algorithm for adjacent frames.
Fig. 6.
Classification examples. Image (a) shows an example of lipid-rich tissue (L); image (b) a calcified plaque (C); image (c) a fibrotic plaque (F); image (d) a mixed plaque containing both calcific and lipid-rich tissues (arrow). From the images it is possible to appreciate automated characterization of the 3 main plaque components and outliers (last row) by comparing to ground truth (manual analysis - middle row). Asterisks indicate guide-wire shadowing artifact and (†) indicates a side-branch. Color coding: yellow = lipid-rich, green = fibrotic, white = calcific and gray = outliers.
Fig. 7.
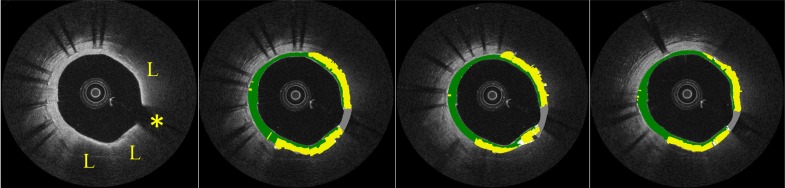
Analysis of 3 adjacent frames. Automated tissue characterization procedure shows consistency through multiple images. Asterisk indicates guide-wire shadowing artifact while (L) indicates lipid-rich tissue.
An average number of ~2.6·104 pixels per image were analyzed resulting in a total of ~1.05·106 testing pixels. The overall classification accuracy was 81.5% with a feasibility of 76.5%. Single class accuracies of 89.5% for fibrotic tissue, 72.1% for calcium and 79.5% for lipid-rich plaques were found. Table 3 reports measured values (mean ± standard deviation) of both optical properties and textural features and statistical significance. Furthermore, according to RF algorithm, the two most important variables for classification were attenuation and mean, respectively. The third variable in order of importance was the information measures of correlation followed by cluster shade, homogeneity, contrast and entropy. The other variables showed of lower importance for classification.
Table 3. Measured Values of Optical Properties and Textural Features for the Different Plaque Typesa.
| Fibrous | Calcified | Lipid | Outliers | |
|---|---|---|---|---|
| µt | 2.3 ± 1.3 mm−1 | 2.0 ± 0.9 mm−1 | 8.8 ± 1.6 mm−1 | ~ |
| Mean | 356 ± 135 | 98 ± 42 | 253 ± 201 | 16 ± 7 |
| Contrast | 122 ± 93 | 11 ± 10 | 89 ± 118 | 0.6 ± 0.7 |
| Cluster shade | 923 ± 2000 | 59 ± 273* | 1667 ± 3312 | 0.5 ± 5* |
| Energy | 0.012 ± 0.001 | 0.045 ± 0.031 | 0.026 ± 0.020 | 0.328 ± 0.172 |
| Entropy | 4.45 ± 0.09 | 3.52 ± 0.56 | 4.02 ± 0.57 | 1.40 ± 0.44 |
| Homogeneity | 0.21 ± 0.05 | 0.44 ± 0.10 | 0.32 ± 0.13 | 0.77 ± 0.07 |
| Inverse difference moment | 0.97 ± 0.02 | 0.70 ± 0.16 | 0.85 ± 0.16 | 0.18 ± 0.11 |
| Information measures of correlation | 0.97 ± 0.01 | 0.99 ± 0.002 | 0.98 ± 0.02 | 1 ± 10−4 |
aValues are reported as mean ± standard deviation. In order to report statistical significance a one-way ANOVA test (and Bonferroni post-hoc testing when appropriate) was applied. All groups have means significantly different from each other with the exception of the two groups labeled with an asterisk (*).
Computational time for the analysis of a sample of 104 pixels was ~30 seconds. More specifically, the time required by the scan-conversion algorithm was ~1 second for an output image of pixel dimensions 2000 by 2000. Single image lumen segmentation and attenuation analysis required ~0.6 and ~1.4 seconds respectively. Computation of the co-occurrence matrix and the textural features required a total of ~27 seconds while Random Forests classification took approximately 2.8 seconds. All reported computational times were quantified on a standard office pc, without any specific optimization.
7. Discussion
In this study, we presented an algorithm for the automated characterization of atherosclerotic tissue on IVOCT images. The algorithm was validated by analyzing images of 40 atherosclerotic plaques both manually and by the proposed methodology showing good results for both overall and per-class accuracy. Given that the test set was extracted from 31 in vivo IVOCT pullbacks, i.e. showing a wide range of image quality and artifacts, this study indicates that the proposed algorithm provides a reliable automated atherosclerotic tissue analysis, able to characterize plaque components in vivo.
7.1 Accuracy
Overall, a classification accuracy of 81.5% was found. This accuracy was obtained in realistic clinical imaging conditions in which image quality may have been suboptimal and different kinds of artifacts can occur such as incomplete vessel flushing, eccentricity of the catheter, tangential signal dropout or motions artifacts [16,40]. Although the accuracy seems acceptable, to the authors’ best knowledge, this is the first attempt to automatically classify the three main plaque components by means of IVOCT in vivo implying that no comparison can be made to prior art.
Analysis of adjacent frames of a randomly selected pullback showed consistent classification results (Fig. 7), indicating that the automated classifier produces consistent results as all frames were processed independently and thus no spatial continuity in the classification result was imposed.
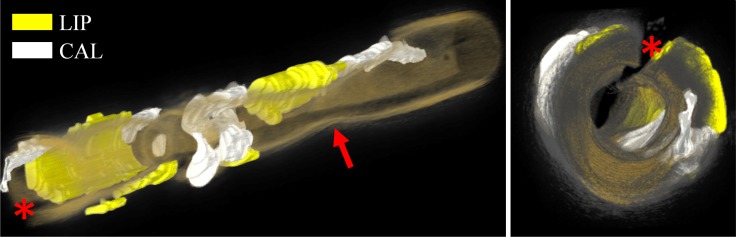
The accuracy and consistency of the proposed algorithm therefore indicates that IVOCT offers, for the first time, the ability to analyze atherosclerotic plaques in vivo with a resolution <20µm in an automated and therefore time efficient manner. As such, it makes processing entire pullbacks feasible, enabling a 3D visualization of the tissue distribution with the vessel wall (Fig. 8) potentially improving diagnostic and therapeutic value of these data sets. However, the maximum penetration depth of this imaging approach remains limited by OCT technology especially in case of high attenuating tissues, implying that its volume may be intrinsically impractical to assess. However, a circumferential quantification of fibrotic, calcified and lipid-rich plaques can be achieved.
Fig. 8.
3D rendering of an entire vessel volume depicting different plaque components. Multiple adjacent IVOCT images are automatically analyzed and further processed with dedicated software (OsiriX [41]). Prior to rendering, results were visually inspected and manually corrected where needed. Images immediately illustrate the 3D distribution of atherosclerotic tissue in the analyzed vessel. The arrow points to a vessel stenosis presenting the IVOCT minimal lumen area. The red asterisk indicates guide-wire shadowing artifact. Color coding: yellow = lipid-rich, white = calcific tissue and orange-to-gray rendering of the vessel wall.
7.2 Classification parameters
Two optical properties of the atherosclerotic tissue components were used for their classification: scattering and attenuation. The former was assessed through texture analysis, i.e. quantifying the stochastic behavior of the backscatter process within a spatially continuous region of interest that depends on the exact spatial distribution and properties of the individual scattering sites, while the latter was quantified by fitting a signal model to experimentally recorded data. These two optical parameters were chosen as they have been identified as properties that experts use to visually classify atherosclerotic tissue on IVOCT images [11,12,16]. According to the results of this study, attenuation and mean pixel intensity (first order statistics) were the parameters with higher importance for atherosclerotic tissue classification. The attenuation coefficient has previously been shown to be quantitatively different between different kinds of atherosclerotic tissue components [12,14,27]. Obviously, texture (i.e. scattering) characteristics further contribute in the classification of plaque components with similar attenuation properties (i.e. fibrotic and calcified tissues).
The proposed signal model used for attenuation estimation assumes the wall to consist of distinct, homogeneous layers in which a single scattering approximation is appropriate. Although more complex signal models can take the effect of multiple scattering into account [42], they will increase computational complexity of the fitting procedure and make the solution more dependent on the initialization settings of the optimization scheme. Moreover, the effect of multiple scattering with respect to attenuation estimation is considered to be very limited. As a matter of fact, single scattering theory is commonly used in literature to describe the OCT signal in biological tissues [14,24,27]. In addition, the signal model used in this study corrects for the catheter PSF but does not take into account the roll-off of the OCT system. Although this effect was reported to be small, adding to Eq. (3) a term for roll-off compensation [14] may further increase attenuation estimation accuracy.
The absolute attenuation coefficient found by this multi-layer model resulted in comparable values with the former studies of van Soest et al. [14] and Xu et al. [12] indicating proper functioning of the proposed methodology. Of note is that occasionally ‘positive’attenuation values could be obtained in the fitting procedure as a result of spatial variations in backscatter coefficient µb, leading to a local increase of signal amplitude with depth. Given that this would result in a non-physical solution, the attenuation coefficient of these regions was set to zero, similar to what was done in previous studies [14]. An alternative solution may consist in the use of additional constrains in the fitting procedure such as extending the layer width in case ‘positive’ attenuation is found. However, this kind of solutions may limit the accuracy of the fitting procedure, as two different tissues may result to be partially incorporated within the same layer.
7.3 Choice of the classifier
It has been shown that, for image classification applications, RF shows comparable performance to other state-of-the-art methods (e.g. multi-way Support Vector Machines) while reducing computational costs [43,44]. As such, RF is often used in case of very large training data sets and a very large number of input variables (e.g. > 103-4) as it was in this study. Moreover, it has been shown that, when the number of trees to grow ntree is large enough, RF does not overfit and is very robust with respect to noise [31]. In addition, RF is inherently multi-class. Because of these characteristics, RF resulted to be an appropriate choice for the problem addressed in this study.
7.4 Study limitations
Validation was performed using manual analysis of an expert image reader as the ground truth. Accuracy was quantified as typically done for computer vision algorithms for object recognition and segmentation [38] and former methods for intravascular ultrasound (IVUS) tissue characterization [7]. Although manual analysis of IVOCT images is prone to at least some inter-operator variability [13], and thus resulting in a weaker ground truth in respect to histology, the analysis was performed by a reader blinded to the results of the algorithm to avoid bias. Moreover, bias was further reduced by applying the algorithm to exactly the same ROIs traced during manual analysis. Anyway, only a single image reader was used as ground truth. Given that IVOCT manual assessment of atherosclerotic plaques is subject to some inter-observer variability, the use of a second image reader could have given more detailed information about the performance of the proposed method. However, especially for a correct training, the use of a third reader is always required in case of disagreement between two readers. As the quantification of intra- and inter-observer variability was the subject of previous publications, it was beyond the scope of this study. As a matter of fact, only training and validation using a large series of histological samples would be able to give more objective and detailed results. However, even if histology can provide a stronger ground truth, the correct registration with IVOCT images can be challenging due to histological slice thickness and helicoidal IVOCT data acquisition [45,46]. Moreover, given the different resolution between the two modalities, a pixels-wise registration cannot be achieved. In addition, a large amount of histological data would be required to achieve enough statistical power, which is not currently available.
Besides these considerations, a possible limitation is that manual analysis typically tends to depict plaques as homogeneous regions of atherosclerotic tissue. This is not necessary true as atherosclerosis is a progressive disease and a plaque of a given type may also contain a limited amount of different tissues. As the classification algorithm performs individual pixel analysis, these inhomogeneities are taken into account by the automated procedure. Thereby, such difference between the two procedures may have reduced the algorithm accuracy results as assessed by our validation study. Nevertheless, it is important to emphasize that, although it is very reasonable to expect some inhomogeneities within an atherosclerotic plaque (e.g. a small area of lipidic tissue within a calcified plaque, Fig. 6(b)), isolated pixels (e.g. a single white pixel within a green area, Fig. 6(c)) should be considered ‘classification noise’ and thus not be interpreted as true tissue inhomogeneities.
7.5 Further improvements
Computational times for scan-conversion, lumen segmentation, attenuation estimation and RF classification algorithms are in the order of seconds (<2 sec). Longer computational times (~30 seconds) were found for the Matlab code for texture analysis. As such, further code optimization and implementation in a faster language (e.g. C/C++ or CUDA) would significantly reduce computational time, possibly achieving the analysis of a single OCT image in a time <1 seconds allowing for a fast analysis of multiple IVOCT images.
A limitation of the proposed algorithm is that the image analysis procedure requires the user input for the selection of ROIs for plaque assessment. However, once the ROI is selected, plaque characterization runs fully-automatic. Ulterior developments of the algorithm for the automatic segmentation of peri-vascular tissue (e.g. texture based segmentation) and discrimination of normal artery wall (e.g. additional geometrical features) would allow for a fully automated assessment of images and thus complete IVOCT pullbacks.
In addition, the use of 2D and 3D spatial continuity (especially for future generations of IVOCT systems with a higher frame aiming to reduce the longitudinal distance between multiple frames) may potentially improve final tissue characterization results, making areas of atherosclerotic tissue classified in a more homogeneous way. Although further developments in this direction may be particularly challenging (e.g. discrimination of what has to be considered ‘classification noise’ or not), this may allow for the classification of atherosclerotic tissue in an even more consistent and robust manner.
Finally, it should be noted that the presence of macrophages was not considered in this study. Previous publications [47] showed that macrophage measurements can be obtained through the use of a feature called normalized standard deviation (NSD). Although this is a very interesting category that can be added to the method presented in this study, it was indicated that NSD must be applied only in the context of fibroatheroma caps as other components, such as internal elastic lamina and some calcification, may also generate an elevated NSD value [47]. As such, this can only be applied as a post-processing step once lipidic plaques are correctly localized. By doing this, automatic macrophage quantification can also be obtained as an extension of the method illustrated in this study. Similarly, the quantification of fibroatheroma cap thickness can be also added as a further post-processing step. However, a correct validation of both macrophage and cap thickness quantification would require the use of histological data which are not currently available.
7.6 Clinical implications and future prospects
Detection and morphological assessment of superficial TCFA, fibrotic and calcified plaques (CP) play an important role in the understanding of atherosclerotic disease and for the development of new drugs and therapies. As an example, coronary plaque composition can have an impact on the outcome of percutaneous coronary interventions (PCI). Stents overlying a lipidic core have been associated with long-term adverse healing responses within the stent [5,15,48,49], while stents implanted over a severely calcified region tend to have a higher risk of underexpansion, associated with higher risk of stent failure (e.g. stent thrombosis and in-stent restenosis) [50]. An automatic method for the characterization and quantification of the disease in a specific coronary artery segment (as shown in Fig. 8) could be valuable both for clinical research and, in a future prospect, in guiding the interventional cardiologist during coronary interventions.
8. Conclusion
We developed an algorithm for the automated atherosclerotic tissue characterization in vivo through IVOCT images. A validation study suggested that the main plaque components can be automatically classified and characterized. This will possibly contribute to the understanding of coronary atherosclerotic disease, aid in the assessment of the effect of new drugs and therapies on plaques, and, more in general, to the use of IVOCT in interventional cardiology research and clinical practice.
Acknowledgments
The research program is partially founded by a grant from the Research Foundation Flanders FWO and by a KU Leuven IOF grant. Dr. Tom Adriaenssens is also supported by a clinical doctoral grant from the FWO foundation.
Appendix
Feature selection consists in the identification of a reduced set of features that can best distinguish between the different types of tissue. Starting from the 24 images composing the training-set, 22 statistical features were quantified and stored. SGLDMs were initially computed in a squared kernel of pixel dimension K=5-7-9-11-13 for every pixel to analyze with the following parameters: distance d=1-2, orientation θ=0°-45°-90°-135° and G=32-64-128 (number of gray levels for quantization). Subsequently, best class separability was assessed by visual inspection of displayed features values against the type of tissue they belong according to previous manual analysis. First, only features presenting an overall acceptable visual separability were retained. Second, single values for parameters G, d, K and θ were arbitrarily selected looking for the minimum set of parameters presenting the best class separability. In addition, to further reduce the set dimensionality, features presenting a Pearson’s correlation index >0.85 were arbitrarily removed. At the end of this procedure, a total of 7 features computed with d=2, θ=0°-90°, K=11 and G=64 were retained. Selected features, as reported on Table 2, resulted to be: (1) contrast, (2) cluster shade, (3) energy, (4) entropy, (5) homogeneity, (6) information measures of correlation and (7) inverse difference moment.
References and links
- 1.Farb A., Burke A. P., Tang A. L., Liang T. Y., Mannan P., Smialek J., Virmani R., “Coronary plaque erosion without rupture into a lipid core. A frequent cause of coronary thrombosis in sudden coronary death,” Circulation 93(7), 1354–1363 (1996). 10.1161/01.CIR.93.7.1354 [DOI] [PubMed] [Google Scholar]
- 2.Virmani R., Burke A. P., Kolodgie F. D., Farb A., “Pathology of the thin-cap fibroatheroma: a type of vulnerable plaque,” J. Interv. Cardiol. 16(3), 267–272 (2003). 10.1034/j.1600-0854.2003.8042.x [DOI] [PubMed] [Google Scholar]
- 3.Serruys P. W., Luijten H. E., Beatt K. J., Geuskens R., de Feyter P. J., van den Brand M., Reiber J. H., ten Katen H. J., van Es G. A., Hugenholtz P. G., “Incidence of restenosis after successful coronary angioplasty: a time-related phenomenon. A quantitative angiographic study in 342 consecutive patients at 1, 2, 3, and 4 months,” Circulation 77(2), 361–371 (1988). 10.1161/01.CIR.77.2.361 [DOI] [PubMed] [Google Scholar]
- 4.Kang S. J., Mintz G. S., Park D. W., Lee S. W., Kim Y. H., Whan Lee C., Han K. H., Kim J. J., Park S. W., Park S. J., “Mechanisms of in-stent restenosis after drug-eluting stent implantation: intravascular ultrasound analysis,” Circ. Cardiovasc. Interv. 4(1), 9–14 (2011). 10.1161/CIRCINTERVENTIONS.110.940320 [DOI] [PubMed] [Google Scholar]
- 5.Papayannis A. C., Abdel-Karim A. R., Mahmood A., Rangan B. V., Makke L. B., Banerjee S., Brilakis E. S., “Association of coronary lipid core plaque with intrastent thrombus formation: a near-infrared spectroscopy and optical coherence tomography study,” Catheter. Cardiovasc. Interv. 81(3), 488–493 (2013). 10.1002/ccd.23389 [DOI] [PubMed] [Google Scholar]
- 6.Moussa I., Di Mario C., Moses J., Reimers B., Di Francesco L., Martini G., Tobis J., Colombo A., “Coronary stenting after rotational atherectomy in calcified and complex lesions. Angiographic and clinical follow-up results,” Circulation 96(1), 128–136 (1997). 10.1161/01.CIR.96.1.128 [DOI] [PubMed] [Google Scholar]
- 7.Nair A., Kuban B. D., Tuzcu E. M., Schoenhagen P., Nissen S. E., Vince D. G., “Coronary plaque classification with intravascular ultrasound radiofrequency data analysis,” Circulation 106(17), 2200–2206 (2002). 10.1161/01.CIR.0000035654.18341.5E [DOI] [PubMed] [Google Scholar]
- 8.Cordeiro M. A S., Lima J. A C., “Atherosclerotic plaque characterization by multidetector row computed tomography angiography,” J. Am. Coll. Cardiol. 47(8Suppl), C40–C47 (2006). 10.1016/j.jacc.2005.09.076 [DOI] [PubMed] [Google Scholar]
- 9.Jang I. K., Bouma B. E., Kang D. H., Park S. J., Park S. W., Seung K. B., Choi K. B., Shishkov M., Schlendorf K., Pomerantsev E., Houser S. L., Aretz H. T., Tearney G. J., “Visualization of coronary atherosclerotic plaques in patients using optical coherence tomography: comparison with intravascular ultrasound,” J. Am. Coll. Cardiol. 39(4), 604–609 (2002). 10.1016/S0735-1097(01)01799-5 [DOI] [PubMed] [Google Scholar]
- 10.Jang I. K., Tearney G. J., MacNeill B., Takano M., Moselewski F., Iftima N., Shishkov M., Houser S., Aretz H. T., Halpern E. F., Bouma B. E., “In vivo characterization of coronary atherosclerotic plaque by use of optical coherence tomography,” Circulation 111(12), 1551–1555 (2005). 10.1161/01.CIR.0000159354.43778.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yabushita H., Bouma B. E., Houser S. L., Aretz H. T., Jang I. K., Schlendorf K. H., Kauffman C. R., Shishkov M., Kang D. H., Halpern E. F., Tearney G. J., “Characterization of human atherosclerosis by optical coherence tomography,” Circulation 106(13), 1640–1645 (2002). 10.1161/01.CIR.0000029927.92825.F6 [DOI] [PubMed] [Google Scholar]
- 12.Xu C., Schmitt J. M., Carlier S. G., Virmani R., “Characterization of atherosclerosis plaques by measuring both backscattering and attenuation coefficients in optical coherence tomography,” J. Biomed. Opt. 13(3), 034003 (2008). 10.1117/1.2927464 [DOI] [PubMed] [Google Scholar]
- 13.Kume T., Akasaka T., Kawamoto T., Watanabe N., Toyota E., Neishi Y., Sukmawan R., Sadahira Y., Yoshida K., “Assessment of coronary arterial plaque by optical coherence tomography,” Am. J. Cardiol. 97(8), 1172–1175 (2006). 10.1016/j.amjcard.2005.11.035 [DOI] [PubMed] [Google Scholar]
- 14.van Soest G., Goderie T., Regar E., Koljenović S., van Leenders G. L., Gonzalo N., van Noorden S., Okamura T., Bouma B. E., Tearney G. J., Oosterhuis J. W., Serruys P. W., van der Steen A. F., “Atherosclerotic tissue characterization in vivo by optical coherence tomography attenuation imaging,” J. Biomed. Opt. 15(1), 011105 (2010). 10.1117/1.3280271 [DOI] [PubMed] [Google Scholar]
- 15.Wang Z., Kyono H., Bezerra H. G., Wang H., Gargesha M., Alraies C., Xu C., Schmitt J. M., Wilson D. L., Costa M. A., Rollins A. M., “Semiautomatic segmentation and quantification of calcified plaques in intracoronary optical coherence tomography images,” J. Biomed. Opt. 15(6), 061711 (2010). 10.1117/1.3506212 [DOI] [PubMed] [Google Scholar]
- 16.Tearney G. J., Regar E., Akasaka T., Adriaenssens T., Barlis P., Bezerra H. G., Bouma B., Bruining N., Cho J.-, Chowdhary S., Costa M. A., de Silva R., Dijkstra J., Di Mario C., Dudeck D., Falk E., Feldman M. D., Fitzgerald P., Garcia H., Gonzalo N., Granada J. F., Guagliumi G., Holm N. R., Honda Y., Ikeno F., Kawasaki M., Kochman J., Koltowski L., Kubo T., Kume T., Kyono H., Lam C. C. S., Lamouche G., Lee D. P., Leon M. B., Maehara A., Manfrini O., Mintz G. S., Mizuno K., Morel M.-, Nadkarni S., Okura H., Otake H., Pietrasik A., Prati F., Räber L., Radu M. D., Rieber J., Riga M., Rollins A., Rosenberg M., Sirbu V., Serruys P. W. J. C., Shimada K., Shinke T., Shite J., Siegel E., Sonada S., Suter M., Takarada S., Tanaka A., Terashima M., Troels T., Uemura S., Ughi G. J., van Beusekom H. M. M., van der Steen A. F. W., van Es G.-A., van Soest G., Virmani R., Waxman S., Weissman N. J., Weisz G., International Working Group for Intravascular Optical Coherence Tomography (IWG-IVOCT) , “Consensus standards for acquisition, measurement, and reporting of intravascular optical coherence tomography studies: a report from the International Working Group for Intravascular Optical Coherence Tomography Standardization and Validation,” J. Am. Coll. Cardiol. 59(12), 1058–1072 (2012). 10.1016/j.jacc.2011.09.079 [DOI] [PubMed] [Google Scholar]
- 17.Gossage K. W., Tkaczyk T. S., Rodriguez J. J., Barton J. K., “Texture analysis of optical coherence tomography images: feasibility for tissue classification,” J. Biomed. Opt. 8(3), 570–575 (2003). 10.1117/1.1577575 [DOI] [PubMed] [Google Scholar]
- 18.Otsu A., “A threshold selection method from gray-level histograms,” IEEE Trans. Syst. Man Cybern. 9(1), 62–66 (1979). 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- 19.E. R. Dougherty, An Introduction to Morphological Image Processing (SPIE Optical Engineering Press, 1992). [Google Scholar]
- 20.Ughi G. J., Adriaenssens T., Onsea K., Kayaert P., Dubois C., Sinnaeve P., Coosemans M., Desmet W., D’hooge J., “Automatic segmentation of in-vivo intra-coronary optical coherence tomography images to assess stent strut apposition and coverage,” Int. J. Cardiovasc. Imaging 28(2), 229–241 (2012). 10.1007/s10554-011-9824-3 [DOI] [PubMed] [Google Scholar]
- 21.Schmitt J. M., Knüttel A., Yadlowsky M., Eckhaus M. A., “Optical-coherence tomography of a dense tissue: statistics of attenuation and backscattering,” Phys. Med. Biol. 39(10), 1705–1720 (1994). 10.1088/0031-9155/39/10/013 [DOI] [PubMed] [Google Scholar]
- 22.Schmitt J. M., Knüttel A., Bonner R. F., “Measurement of optical properties of biological tissues by low-coherence reflectometry,” Appl. Opt. 32(30), 6032–6042 (1993). 10.1364/AO.32.006032 [DOI] [PubMed] [Google Scholar]
- 23.V. Tuchin, Tissue Optics (SPIE Optical Engineering Press, 2007). [Google Scholar]
- 24.van der Meer F. J., Faber D. J., Sassoon D. M. B., Aalders M. C., Pasterkamp G., van Leeuwen T. G., “Localized measurement of optical attenuation coefficients of atherosclerotic plaque constituents by quantitative optical coherence tomography,” IEEE Trans. Med. Imaging 24(10), 1369–1376 (2005). 10.1109/TMI.2005.854297 [DOI] [PubMed] [Google Scholar]
- 25.van Leeuwen T. G., Faber D. J., Aalders M. C., “Measurement of the axial point spread function in scattering media using single-mode fiber based optical coherence tomography,” IEEE J. Sel. Top. Quantum Electron. 9(2), 227–233 (2003). 10.1109/JSTQE.2003.813299 [DOI] [Google Scholar]
- 26.S. L. Jacques, D. Levitz, R. Samatham, D. S. Gareau, N. Choudhury, and F. Truffer, Light Scattering in Confocal Reflectance Microscopy (McGraw-Hill, 2009). [Google Scholar]
- 27.van der Meer F. J., Faber D. J., Perrée J., Pasterkamp G., Baraznji Sassoon D., van Leeuwen T. G., “Quantitative optical coherence tomography of arterial wall components,” Lasers Med. Sci. 20(1), 45–51 (2005). 10.1007/s10103-005-0336-z [DOI] [PubMed] [Google Scholar]
- 28.Haralick A., Shanmugam K., Dinstein I., “Texture features for image classification,” IEEE Trans. Syst. Man Cybern. 3(6), 610–621 (1973). 10.1109/TSMC.1973.4309314 [DOI] [Google Scholar]
- 29.Conners R. W., Harlow C. A., “A theoretical comparison of texture algorithms,” IEEE Trans. Pattern Anal. Mach. Intell. PAMI-2(3), 204–222 (1980). 10.1109/TPAMI.1980.4767008 [DOI] [PubMed] [Google Scholar]
- 30.Clausi D. A., “An analysis of co-occurence texture statistics as a function of gray level quantization,” Can. J. Rem. Sens. 28(1), 45–62 (2002). 10.5589/m02-004 [DOI] [Google Scholar]
- 31.Breiman L., “Random Forests,” Mach. Learn. 45(1), 5–32 (2001). 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 32.Biau G., Devroye L., Lugosi G., “Consistency of Random Forests and other averaging classifiers,” J. Mach. Learn. Res. 9(1), 2015–2033 (2008). [Google Scholar]
- 33.L. Breiman, “Consistency for a simple model of random forests,” UC Berkeley, Technical Report http://www.stat.berkeley.edu/~breiman/RandomForests/consistencyRFA.pdf (2004).
- 34.Lin Y., Jeon Y., “Random Forests and adaptive nearest neighbours,” J. Am. Stat. Assoc. 101(474), 578–590 (2006). 10.1198/016214505000001230 [DOI] [Google Scholar]
- 35.Ughi G. J., Adriaenssens T., Larsson M., Dubois C., Sinnaeve P. R., Coosemans M., Desmet W., D’hooge J., “Automatic three-dimensional registration of intravascular optical coherence tomography images,” J. Biomed. Opt. 17(2), 026005 (2012). 10.1117/1.JBO.17.2.026005 [DOI] [PubMed] [Google Scholar]
- 36.W. S. Rasband, “ImageJ,” U. S. National Institutes of Health, http://imagej.nih.gov/ij/, Bethesda, Maryland, USA, 1997–2011.
- 37.Abramoff M. D., Magalhales P. J., Ram S. J., “Image processing with ImageJ,” Biophotonics Int. 11(7), 36–42 (2004). [Google Scholar]
- 38.Shotton J., Winn J., Rother C., Criminisi A., “TextoonBoost for image understanding: multi-class object recognition and segmentation by jointly modeling texture, layout and contextq,” Int. J. Comput. Vis. 81(1), 2–23 (2009). 10.1007/s11263-007-0109-1 [DOI] [Google Scholar]
- 39.Metz C. E., “Basic principles of ROC analysis,” Semin. Nucl. Med. 8(4), 283–298 (1978). 10.1016/S0001-2998(78)80014-2 [DOI] [PubMed] [Google Scholar]
- 40.van Soest G., Regar E., Goderie T. P., Gonzalo N., Koljenović S., van Leenders G. J., Serruys P. W., van der Steen A. F., “Pitfalls in plaque characterization by OCT: image artifacts in native coronary arteries,” JACC Cardiovasc. Imaging 4(7), 810–813 (2011). 10.1016/j.jcmg.2011.01.022 [DOI] [PubMed] [Google Scholar]
- 41.Rosset A., Spadola L., Ratib O., “OsiriX: an open-source software for navigating in multidimensional DICOM images,” J. Digit. Imaging 17(3), 205–216 (2004). 10.1007/s10278-004-1014-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thrane L., Frosz M. H., Jørgensen T. M., Tycho A., Yura H. T., Andersen P. E., “Extraction of optical scattering parameters and attenuation compensation in optical coherence tomography images of multilayered tissue structures,” Opt. Lett. 29(14), 1641–1643 (2004). 10.1364/OL.29.001641 [DOI] [PubMed] [Google Scholar]
- 43.Gislason P. O., Benediktsson J. A., Sveinsson J. R., “Random forests for land cover classification,” Pattern Recognit. Lett. 27(4), 294–300 (2006). 10.1016/j.patrec.2005.08.011 [DOI] [Google Scholar]
- 44.Bosch A., Zisserman A., Munoz X., “Image classification using Random Forests and Ferns,” IEEE 11th Int. Conf. Computer Vision (2007). 10.1109/ICCV.2007.4409066 [DOI] [Google Scholar]
- 45.Rieber J., Meissner O., Babaryka G., Reim S., Oswald M., Koenig A., Schiele T. M., Shapiro M., Theisen K., Reiser M. F., Klauss V., Hoffmann U., “Diagnostic accuracy of optical coherence tomography and intravascular ultrasound for the detection and characterization of atherosclerotic plaque composition in ex-vivo coronary specimens: a comparison with histology,” Coron. Artery Dis. 17(5), 425–430 (2006). 10.1097/00019501-200608000-00005 [DOI] [PubMed] [Google Scholar]
- 46.Meissner O. A., Rieber J., Babaryka G., Oswald M., Reim S., Siebert U., Redel T., Reiser M., Mueller-Lisse U., “Intravascular optical coherence tomography: comparison with histopathology in atherosclerotic peripheral artery specimens,” J. Vasc. Interv. Radiol. 17(2), 343–349 (2006). 10.1097/01.RVI.0000195324.52104.00 [DOI] [PubMed] [Google Scholar]
- 47.Tanaka A., Tearney G. J., Bouma B. E., “Challenges on the frontier of intracoronary imaging: atherosclerotic plaque macrophage measurement by optical coherence tomography,” J. Biomed. Opt. 15(1), 011104 (2010). 10.1117/1.3290810 [DOI] [PubMed] [Google Scholar]
- 48.Nakazawa G., Finn A., Virmani R., “Morphologic predictors of drug-eluting stent thrombosis,” U.S. Cardiol. 4(1), 75–76 (2006). [Google Scholar]
- 49.Imola F., Occhipinti M., Biondi-Zoccai G., Di Vito L., Ramazzotti V., Manzoli A., Pappalardo A., Cremonesi A., Albertucci M., Prati F., “Association between proximal stent edge positioning on atherosclerotic plaques containing lipid pools and postprocedural myocardial infarction (from the CLI-POOL Study),” Am. J. Cardiol. 111(4), 526–531 (2013). 10.1016/j.amjcard.2012.10.033 [DOI] [PubMed] [Google Scholar]
- 50.Kawaguchi R., Tsurugaya H., Hoshizaki H., Toyama T., Oshima S., Taniguchi K., “Impact of lesion calcification on clinical and angiographic outcome after sirolimus-eluting stent implantation in real-world patients,” Cardiovasc. Revasc. Med. 9(1), 2–8 (2008). 10.1016/j.carrev.2007.07.004 [DOI] [PubMed] [Google Scholar]