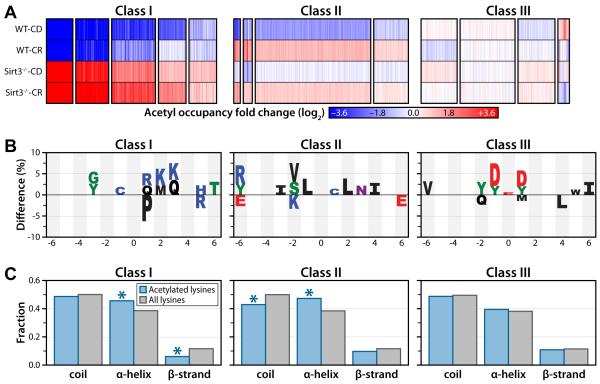
Figure 5. Cluster analysis resolves acetyl sites into three distinct classes.
(A) Cluster analysis identified 15 clusters, with 13 grouped into 3 classes based on their overall trends between the four biological conditions: Class I - Acetylation sites that respond dynamically to SIRT3 expression. Class II - Acetylation sites that are largely independent of SIRT3 expression but respond to CR. Class III - Acetylation sites that display minimal perturbation in response to either loss of SIRT3 expression or CR. Each column represents a quantified acetyl site. Blue: decreased acetyl occupancy, White: no change, Red: increased acetyl occupancy.
(B) Amino acid motifs identified for clusters displaying the largest magnitude fold change for each class (from A). The motif shows significant (p < 0.05) under or over representation of a particular amino acid at each site flanking the acetylated lysine (position 0).
(C) Predicted protein secondary structures of acetylation sites identified in each class. Probabilities for coil, α-helix and β-strands were compared with the probabilities of these secondary structure elements in all lysines identified in the study. (Class I α-helix: p < 3.0 × 10−3; Class I β-strand: p < 2.0 × 10−4; Class II coil: p < 8.0 × 10−4; Class II α-helix: p < 2.5 × 10−5, hyper-geometric analysis).