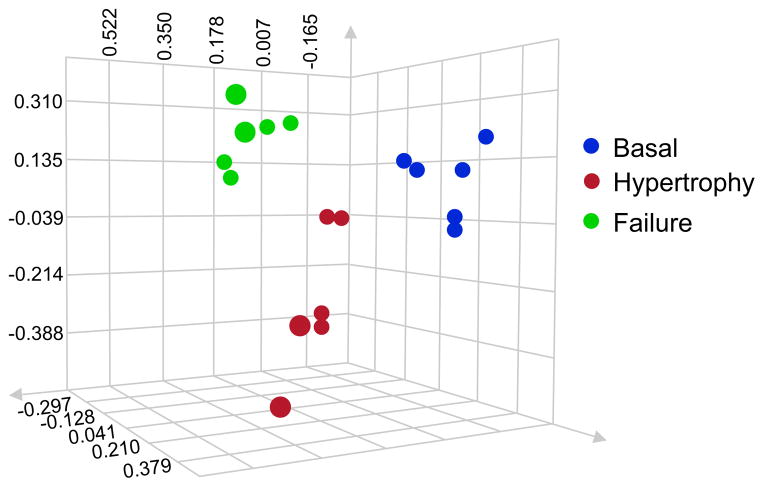
Figure 4. Analysis of variance and principal component analysis.
Cardiac chromatin fractions from 3 basal mice (blue), 3 mice in cardiac hypertrophy (red) and 3 mice in cardiac failure (green) were run on a one-dimensional protein gel. Proteins of 75–150kDa were cut and digested. Each sample was analyzed twice by LC/MS/MS for a technical replicate. The data was searched and protein abundance determined by label-free quantitation. ANOVA analysis was used to generate a PCA plot where axes represent peptide intensity and each dot represents a single replicate. Clustering by disease state reveals distinct proteomic changes occuring during cardiac hypertrophy and failure, and confirms the ability of this methodology to reproducibly detect these changes.