Table 1. Model parameters used in simulations.
| Symbol | value | unit | Comment | |
| First-order rate constant for ADP-ribosylation | k cat | 0.0045 | s−1 | assumed1 |
| Total EF2 in the system | [EF2]t | 0.6 | µM/cell | [36] |
| Total ribosomes engaging in mRNA translation | [R]t | 0.5 | µM/cell | [37] 2 |
| EF1-initiated peptidyl insertion rate constant | k 0,max | 4 | s−1 | [38] |
| Association rate constant between E GTP and R • | k 1 | 96 | µM−1 s−1 | [8], [9] 3 |
Dissociation rate constant for 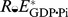
|
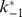
|
0.3 | s−1 | assumed |
Translocation rate constant for 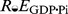
|
k 2 | 35 | s−1 | [8], [9] |
EF2•GDP release rate constant for 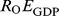
|
k 3 | 5 | s−1 | [8], [9] 4 |
| GTP exchange rate constant for E GDP | k 4 | 12.8 | µM s−1 | [39] |
| EF2•GDP dissociation constant | K d,GDP | 0.5 | µM | [24] |
| EF2•GTP dissociation constant | K d,GTP | 2.68 | µM | [24] |
| Free GTP concentration | [GTP] | 400 | µM | [35] |
| Free GDP concentration | [GDP] | 40 | µM | assumed5 |
| Polysomal density | m | 10 | # ORF−1 | assumed |
| Threshold for dead-end translocation block in f | f c | 0.09 | [17] | |
| Basal fraction of translocation-active ribosomes |
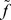
|
0.29 | modeled | |
| Basal stalling coefficient |
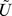
|
0.14 | modeled |
The rate constant 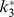 is set equal to k
3 throughout the work, whereas
is set equal to k
3 throughout the work, whereas 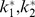 , and
, and 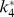 may differ from their native counterparts.
may differ from their native counterparts.
Calculated from λ cat [T], using λ cat = 1.7×109 M−1 min−1 [34] and [T] = 0.16 nM (equivalent to 250 toxin molecules in the cytosol of a cell, with a cell cytosol volume equal to 2.6 pL [35].
Obtained by the ratio of [EF2]t : [R]t = 1.2∶ 1 [37] using the estimated [EF2]t.
Calculated as k 1 = k a k GTP/(k d+k GTP) = 150*250/(140+250) = 96 s−1.
Based on [GTP]:[GDP] = 10∶1 [32].
