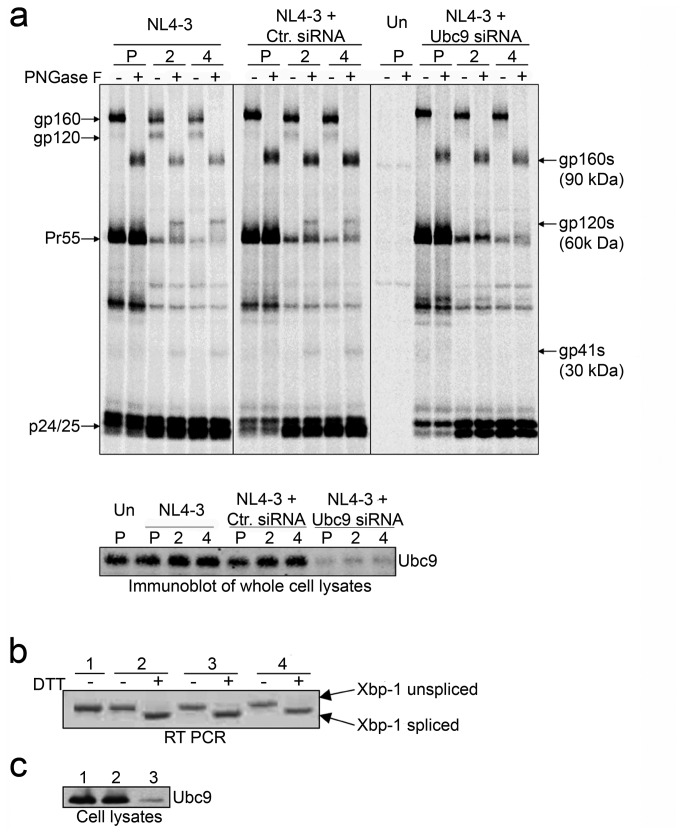
Figure 1. Decreased Env stability in Ubc9 knockdown cells is not due to endoplasmic reticulum stress responses.
(a) Molecular weights of deglycosylated Env proteins in the presence or knockdown of Ubc9 expression. 293T cells were transfected with pNL4-3 alone, or in combination with either Ctr. siRNA or Ubc9 siRNA, or left untransfected (Un). Cells were pulse (P) labeled with [35S] methionine/cysteine for 1 hour and chased for 2 and 4 hours. Cell associated viral proteins were solublized and immunoprecipitated with pooled AIDS patient sera, split equally, and incubated for 3.5 hours at 37°C in the presence, or absence of N-glycosidase F (PNGase F). Samples were separated by SDS PAGE and visualized by phosphorimaging using The Discovery Series Quantity One software. A representative over-exposed gel is shown so that partially Endo Hf resistant Env can be more easily visualized. The identity and position of PNGase F untreated viral proteins are labeled on the left of the gel. Deglycosylated, PNGase F sensitive, viral proteins are labeled on the right and are denoted with a (s) to identify the position of gp160s, gp120s, and gp41s in the gel after PNGase F treatment. (b) Unfolded protein response activation state in the presence or knockdown of Ubc9 expression. Untransfected (1), NL4-3 alone (2), NL4-3 + Ctr. siRNA (3), NL4-3 + Ubc9 siRNA (4). Total RNA was extracted from transfected 293t cells that were either treated with 5mM DTT for 3 hours to induce UPR, or left untreated. Total mRNA was reverse transcribed with oligo (dT) followed by PCR amplification of XBP-1 cDNA using primers that flank an alternative splicing site with in the XBP-1 mRNA. (c) Immunoblot of Ubc9 expression in whole cell lysates. Untransfected (group 1), Ctr. siRNA (group 2), and Ubc9 siRNA (group 3).