Figure 1.
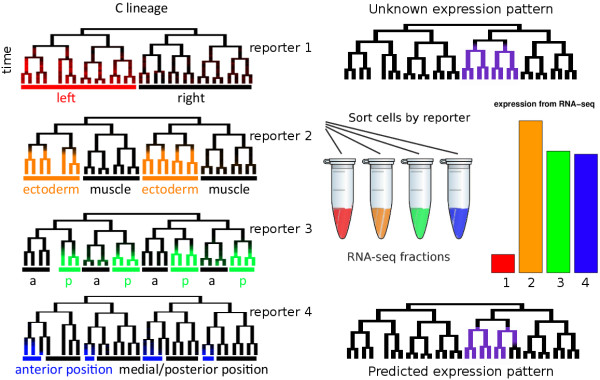
Illustration of the method. We assume that we know the expression patterns of a set of reporters (subset of four reporters expression across ~31 terminal cells and their ancestors shown on the left – the full dataset annotates the expression of 127 reporters across all cells). Each expression pattern is drawn superimposed on a lineage tree. These trees show a group of related cells from the C. elegans lineage with divisions denoted by bifurcations on the on the x axis and time on the y axis. Because of the invariant development, each embryo expressing a given reporter always has reporter expression in the same cells on the lineage, and this is a perfect proxy for cell fate and position. We then flow-sort cells which are expressing each reporter, and perform RNA-seq on the resulting fractions of cells. Based on these measurements, we attempt to estimate expression of each gene in each cell.
