Figure 1. Example of a complex deletion generated by FoSTeS/MMBIR and a pipeline for predicting SV mechanisms.
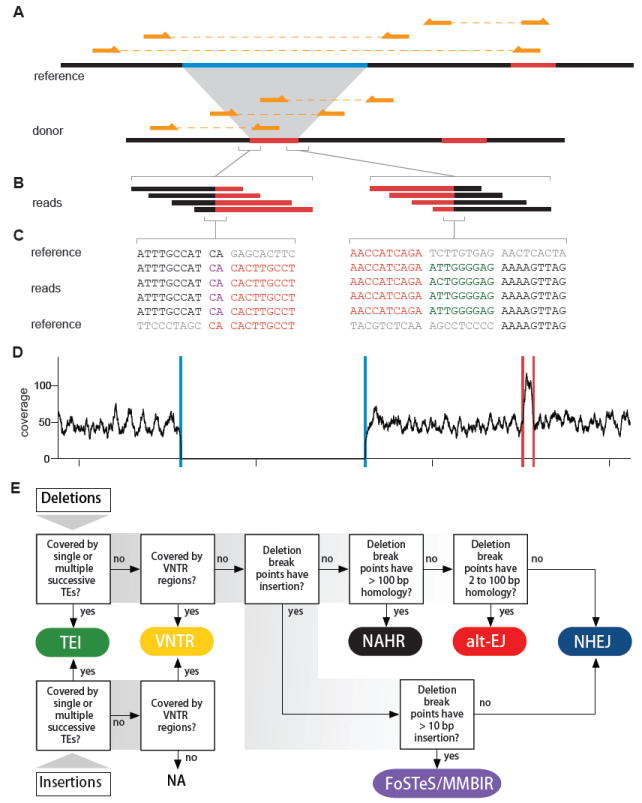
(A) A complex deletion is predicted by three discordant clusters. The sequence in light blue on the reference is deleted; the sequence in red on the reference is duplicated and inserted into the deletion breakpoints. Three read pairs from the donor are shown above the donor sequence. Three discordant read pairs mapped to the reference are shown above the reference sequence.
(B) Reads covering the breakpoints of insertion. The breakpoints are covered by 27 and 11 reads, respectively (only four are shown for each). Reads matching different parts of the reference genome are shown in the corresponding colors.
(C) Nucleotide sequences of the reads covering the breakpoints of insertion. Black and red colors indicate the reads and the reference sequences that match each other and the grey sequences indicate unmatched references. There are a 2 bp microhomology (shown in purple) at the breakpoint on the left and a 9 bp insertion of unknown source (shown in dark green) at the breakpoint on the right.
(D) Sequencing depth. Blue and red lines denote the predicted deletion and the predicted insertion donor sites, respectively, showing that the copy number is consistent with the SV call.
(E) This flowchart, adapted mainly from Kidd et al. 2010, shows the breakpoint features for determining the mechanism that is likely to have generated the observed SV. Six types of mechanisms are assigned: transposable element insertion (TEI), variable number of tandem repeats (VNTR), non-homologous end joining (NHEJ), alternative end joining (alt-EJ), non-allelic homologous recombination (NAHR) and fork stalling and template switching/microhomology mediated break induced repair (FoSTeS/MMBIR).
See also Figure S1 and Table S1 and S2.
