Abstract
Today, most medical images are stored as a set of single-frame composite Digital Imaging and Communications in Medicine (DICOM) objects that contain the four levels of the DICOM information model—patient, study, series, and instance. Although DICOM addresses most of the issues related to medical image archiving, it has some limitations. Replicating the header information with each DICOM object increases the study size and the parsing overhead. Multi-frame DICOM (MFD) was developed to address this, among other issues. The MFD combines all DICOM objects belonging to a series into a single DICOM object. Hence, the series-level attributes are normalized, and the amount of header data repetition is reduced. In this paper, multi-series DICOM (MSD) is introduced as a potential extension to the DICOM standard that allows faster parsing, transmission, and storage of studies. MSD extends the MFD de-duplication of series-level attributes to study-level attributes. A single DICOM object that stores the whole study is proposed. An efficient algorithm, called the one-pass de-duplication algorithm, was developed to find and eliminate the replicated data elements within the study. A group of experiments were done that evaluate MSD and the one-pass de-duplication algorithm performance. The experiments show that MSD significantly reduces the amount of data repetition and decreases the time required to read and parse DICOM studies. MSD is one possible solution that addresses the DICOM limitations regarding header information repetition.
Keywords: DICOM, Algorithms, Imaging informatics, Image data, Multi-frame DICOM, Multi-series DICOM
Introduction
The Digital Imaging and Communications in Medicine (DICOM) [1] information model arranges the data in four levels: patient, study, series, and instance. Each medical image is stored as a DICOM composite object that contains the four levels of the information model as data elements. While the DICOM standard has enabled strong interoperability between medical imaging devices and applications, it does have some areas where it can be improved. In particular, DICOM communication is still primarily based on single-frame composite objects, in which the patient, study, and series information is replicated in each object. These composite objects are transmitted one object at a time. In addition to duplication, this repetition necessitates parsing and validating the data each time it is transmitted. In typical implementations, which use composite objects, the sending and the receiving application entities have to handshake for each transmitted object. Hence, the time required to transmit the data between different application entities is increased. As the number and size of medical imaging studies continue to increase, finding a solution for these inefficiencies is becoming increasingly important. It should be noted that multi-frame DICOM (MFD) objects are improving this situation by allowing an entire series to be sent as one object; however, there are two problems with the current MFD format: (1) many vendors have not implemented it, and (2) there is no standard way to convert older studies into these objects.1
In this paper, a multi-series DICOM (MSD) object is proposed as a study-level extension of the MFD object. The MFD format stores multiple frames within the same series in one DICOM object in contrast to the DICOM composite object that stores only a single frame. The proposed format stores the whole study in a single MSD object, where each series is potentially a MFD object. The patient, study, and series attributes are stored only once, thus eliminating the repetition of the metadata that occurs with single-frame DICOM objects. In addition, a new algorithm called the one-pass de-duplication algorithm was developed that finds and eliminates the repeated attributes within the traditional single-frame objects. The algorithm eliminates duplicate data elements by allocating them to their appropriate level in the information model. Multiple DICOM studies were used to test the proposed methods. For the test studies, on average, MSD results in metadata that have six times fewer data elements and five times fewer bytes compared to the original single-frame DICOM (SFD). This reduction in the number of data elements results in about 30 % reduction in the time required for reading and parsing the study. Finally, a performance analysis of the one-pass normalization algorithm shows that the process does not add any overhead to the time required to read DICOM studies. This is because the time needed for de-duplication is offset by the cost reduction of not storing duplicate data elements.
Materials and Methods
DICOM supports a nested dataset capability. It is possible to define a sequence element that contains sets of elements. A data element is a sequence data element if its VR value equals to SQ. MFD objects use this capability to store multiple frames belonging to the same series in a single object. The sequence element used to store the frames within the series has a special tag, called the PerFrameFunctionalGroupsSequence tag. This sequence element holds the frame objects. This work extends this idea to the study level and allows storing a whole study in a single MSD object. In order to do this, we introduce a new private tag, called the PerSeriesFunctionalGroupsSequence tag. The proposed MSD object has a special sequence data element with the PerSeriesFunctionalGroupsSequence tag that holds a set of objects where each object represents a different series. Each object representing a series can be a MFD object if the series has more than one frame.
The arrangement of pixel data within the MFD and the MSD objects is important. The MFD objects concatenate the pixel data of all frames within the series and store it in a single-pixel data element. This format requires all frames within the series to have the same size. If a series has frames with different sizes, it cannot be stored in MFD format. In this paper, this issue was addressed using two different approaches. The first approach stores the whole series as one DICOM object. It stores the pixel data of each frame as a pixel data element within the frame's object, which is stored in the PerFrameFunctionalGroupsSequence data element. The second approach creates a MFD object for each possible frame size. Each such object contains a data element with the concatenated pixel data for the frames of that size. The later solution is similar to the MFD standard style of arranging pixel data within a DICOM object, but it creates separate objects for each possible frame size.
An application programming interface (API) was developed to support the MSD object. The API was developed in Java. The implementation was created using the dcm4che2 [http://www.dcm4che.org/confluence/display/d2/dcm4che2+DICOM+Toolkit (accessed January 2012)] toolkit, which is used for reading/writing the DICOM files, as well as parsing and storing the data elements. The API contains a study model that stores the data elements. The study model corresponds to the DICOM information model, i.e., patient, study, series, and instance. In addition, the API includes the de-duplication algorithm that was developed to eliminate duplicate data elements in the study attributes. Finally, the API has an input/output package that reads SFD, MFD, and MSD formats into the study model and allows SFD, MFD, or MSD formats to be written from the study model. The following sections briefly describe the study model, the de-duplication algorithm, and the input/output package.
Study Model
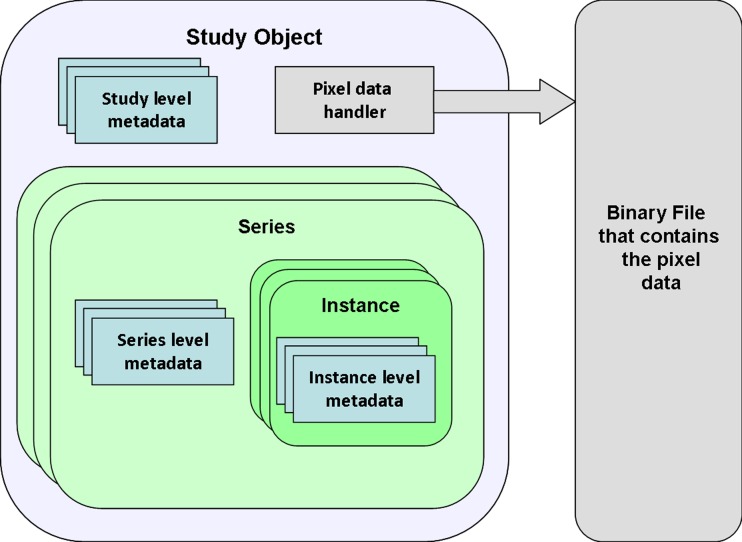
The study model has the same levels of information as the DICOM information model, i.e., study, series, and instance classes. A study object and a binary file represent the DICOM study. The study object is composed of the study-level attributes that are shared among all series and a list of series objects. Each series object is composed of the series-level attributes that are shared among all instances and a list of instances objects. Each instance is composed of the instance-level attributes. The pixel data and other large size attributes (larger than 256 bytes) are stored in the binary file to reduce the memory consumption. The binary file is accessible through the study object; see Fig. 1.
Fig. 1.
Study data model
The One-Pass De-duplication Algorithm
The one-pass de-duplication algorithm constructs the study model described in the previous section. The algorithm finds duplicate attributes within the input study service–object pair (SOP) instances and ensures that the constructed study model is free of repetition. The algorithm depends on the order of the element tags. The DICOM standard requires that all data elements within a SOP instance to be sorted in ascending order according to their tag numbers. The algorithm keeps the study-level elements and series-level elements in order, which enhances performance as illustrated below.
The data elements in the first input DICOM SOP instance are added to the study-level attributes except for the seriesInstanceUID and SOPInstanceUID, which are added to the series-level and instance-level attributes, respectively. For subsequent instances, the algorithm starts with study-level normalization followed by series-level normalization if required.
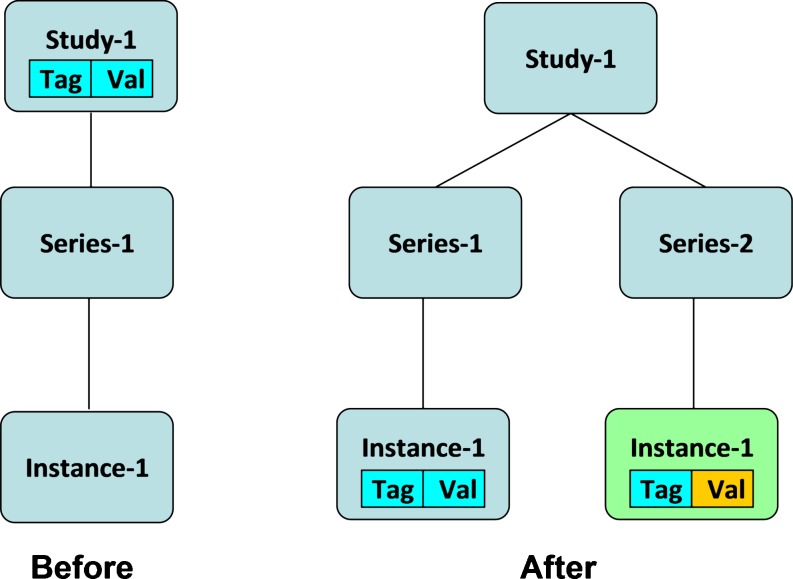
In study-level normalization, the algorithm compares the tag number of the attribute at index inputIndex in the input instance with the tag number of the attribute at studyIndex in the list of study-level attributes. Initially, inputIndex and studyIndex have a value of zero. If the tag numbers are equal and the attribute values are identical, the input attribute is discarded. Both the inputIndex and studyIndex are incremented by one. If the tag numbers are equal but the attributes values are different, the study-level attribute is removed from the list of study-level attributes and added to the list of series-level attributes of all the other series within the study except the series that the input instance belongs to. If the input instance belongs to a new series, the input attribute is added to the list of the series-level attributes of the new series; see Fig. 2.
Fig. 2.
Adding an instance to a study with an attribute that already exists in the study-level attributes but with a different value field. The input instance belongs to a new series
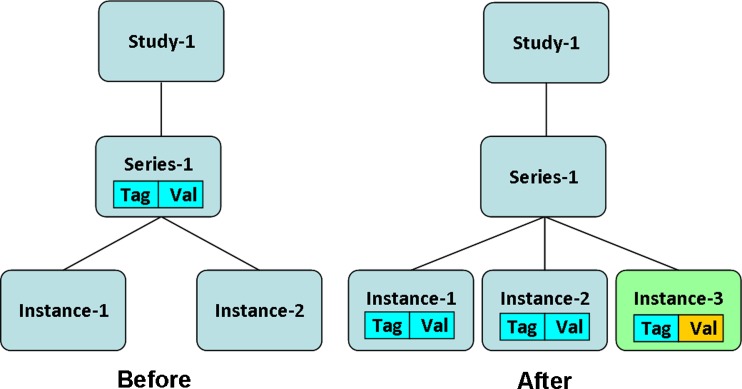
If the input instance belongs to a series that has other instances in it, the input attribute is added to the list of instance-level attribute of the input instance, and the study-level attribute is added to the list of instance-level attributes of each other instance within that series; see Fig. 3. In both cases, the inputIndex and studyIndex are incremented by one.
Fig. 3.
Adding an instance to a study with an attribute that already exists in the study-level attributes but with a different value field. The input instance belongs to a series that has some other instances
If the tag number of the input attribute is greater than the tag number of the study attribute, this implies that the study has a data element that does not exist in the new DICOM SOP instance. Based on this, the attribute is removed from the list of study-level attributes and added to the list of series-level attributes of each series within the study except for the series that the input SOP instance belongs to. If there is only one series, the attribute is added to the list of instance-level attributes of all instances within the series except for the instance that corresponds to the input SOP instance. Finally, the studyIndex is incremented by one.
If the tag number of the input attribute is smaller than the tag number of the study attribute, this implies that the list of study-level attributes does not have an attribute with the same tag. In this case, series-level normalization is applied. Series-level normalization compares the input attribute with the series-level attribute at index seriesIndex. Initially, seriesIndex is zero.
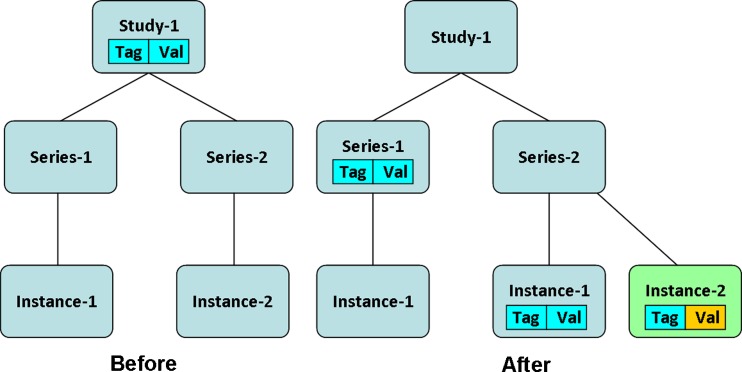
If the tag numbers are equal and the attributes values are identical, the input DICOM attribute is discarded. Both the inputIndex and seriesIndex are incremented by one. If the tag numbers are equal and the attributes' values are different, the input attribute is added to its instance list of instance-level attributes while the series-level attribute is removed from the list of series-level attributes and added to each other instance within the series. Both the inputIndex and seriesIndex are incremented by one; see Fig. 4.
Fig. 4.
Adding an instance to a series with an attribute that already exists in the series-level attributes but with a different value field
If the tag number of the input attribute is greater than the tag number of the series attribute, then the series has a data element that does not exist in the new SOP instance. Based on this, the series-level attribute is removed from the series-level attributes and added to each other instance within the series except for the series that the input object belongs to. The seriesIndex is incremented by one.
If the tag number of the input data element is smaller than the tag number of the series attribute at index seriesIndex, then the list of series-level attributes does not have an attribute with the same tag. The input data element is added to the list of instance-level attributes of its instance, and the inputIndex is incremented by one. The process continues until the inputIndex exceeds the number of attributes in the input SOP instance. This indicates the end of the input SOP instance. The algorithm repeats the same process for each SOP instance within the study.
The Input/Output Package
The input/output (I/O) package supports reading and writing the study model to SFD, MFD, or MSD file formats, see Fig. 5. The output process is simple. For SFD output, each instance in the study model is converted to a DICOM object. The study-level attributes and series-level attributes are added to each SFD object created. For MFD, each series is converted to a DICOM object. The study-level attributes are added to each MFD object created. Each MFD object contains an attribute with PerFrameFunctionalGroupsSequence tag. Instances within the series are converted to DICOM objects stored within PerFrameFunctionalGroupsSequence attribute. Finally, the MSD format stores the whole study in one object where each series is stored as a DICOM object stored in the PerSeriesFunctionalGroupsSequence attribute as described previously.
Fig. 5.
Conversion of SFD, MFD, and MSD formats to/from the study model
A suite of integration tests is used in the development process to validate the conversion process. The tests read studies in SFD, MFD, and SFD formats and output studies in all three file formats. These tests verify that the input study is identical to the output study.
Results
A number of experiments were done to test our proposed MSD object compared to the single-frame DICOM and MFD objects in addition to experiments that evaluate the one-pass de-duplication algorithm performance. For each of the experiments described below, the experiment was run ten times. The outlier runs were thrown away, and the average of the remaining consistent runs was reported. Six different input single-frame DICOM studies were used for our experiments. The size of the input studies ranges between 72 MB and 1.4 GB. For a complete description of the input data set, see Table 1.
Table 1.
Input data sets properties
| Study name | No. of series | No. of frames | Study size (KB) | Binary size (KB) | Header size (KB) |
|---|---|---|---|---|---|
| SMALLMR | 9 | 277 | 72,457 | 71,488 | 969 |
| SMALLCT | 5 | 338 | 174,008 | 173,088 | 920 |
| TESTMR | 17 | 1,116 | 217,630 | 213,352 | 4,278 |
| TESTCT | 7 | 1,018 | 617,236 | 613,926 | 3,310 |
| TESTCTA | 13 | 2,524 | 1,374,373 | 1,366,321 | 8,052 |
| BREASTMR | 22 | 2,362 | 1,508,194 | 1,499,589 | 8,605 |
| Average | 12 | 1,273 | 660,650 | 656,294 | 4,356 |
The first experiment was designed to measure the reduction in the number of data elements and consequently the total size of the metadata after de-duplication. The input studies were saved to a secondary storage device using three different formats, SFD, MFD, and the proposed MSD format. Each of the MFD and MSD formats has two different versions, the first version (V1) with a single-pixel data element per frame size per series and the second version (V2) with a single-pixel data element per frame in the series regardless of its size. Table 2 compares the number of data elements while Table 3 shows the size of the metadata for each format.
Table 2.
Number of DICOM attributes for the DICOM datasets listed in Table 1 stored in different formats. The second column is for SFD format, and the third and fifth are for the MFD and MSD in V1 format. The seventh and ninth columns are for the MFD and MSD in version 2 format. The seventh and ninth columns are for the MFD and MSD in version 2 format. Columns 4 and 8 show the percentages of MFD to SFD while columns 6 and 10 show the percentage of MSD to MFD
| Study name | SFD | MFD (V1) | Percent | MSD (V1) | Percent | MFD (V2) | Percent | MSD (V2) | Percent |
|---|---|---|---|---|---|---|---|---|---|
| SMALLMR | 31,241 | 3,391 | 11 % | 2,695 | 9 % | 3,659 | 12 % | 2,963 | 9 % |
| SMALLCT | 28,777 | 3,753 | 13 % | 3,575 | 12 % | 4,086 | 14 % | 3,908 | 14 % |
| TESTMR | 129,705 | 12,231 | 9 % | 11,049 | 9 % | 13,330 | 10 % | 12,148 | 9 % |
| TESTCT | 105,540 | 35,429 | 34 % | 27,972 | 27 % | 36,235 | 34 % | 28,778 | 27 % |
| TESTCTA | 269,665 | 86,488 | 32 % | 71,303 | 26 % | 88,526 | 33 % | 73,341 | 27 % |
| BREASTMR | 268,149 | 27,955 | 10 % | 26,380 | 10 % | 30,295 | 11 % | 28,720 | 11 % |
| Average | 138,846 | 28,208 | 18 % | 23,829 | 15 % | 29,355 | 19 % | 24,976 | 16 % |
Table 3.
Metadata size of the DICOM datasets listed in Table 1 stored in different formats. The second column is for SFD format, and the third and fifth are for the MFD and MSD in V1 format. The seventh and ninth columns are for the MFD and MSD in version 2 format. Columns 4 and 8 show the percentages of MFD to SFD while columns 6 and 10 show the percentage of MSD to MFD
| Study name | SFD | MFD (V1) | Percent | MSD (V1) | Percent | MFD (V2) | Percent | MSD (V2) | Percent |
|---|---|---|---|---|---|---|---|---|---|
| SMALLMR | 969 | 156 | 16 % | 139 | 14 % | 158 | 16 % | 141 | 15 % |
| SMALLCT | 920 | 171 | 19 % | 166 | 18 % | 174 | 19 % | 169 | 18 % |
| TESTMR | 4,278 | 585 | 14 % | 550 | 13 % | 594 | 14 % | 559 | 13 % |
| TESTCT | 3,310 | 1,193 | 36 % | 890 | 27 % | 1,193 | 36 % | 896 | 27 % |
| TESTCTA | 8,052 | 2,829 | 35 % | 2,201 | 27 % | 2,845 | 35 % | 2,217 | 28 % |
| BREASTMR | 8,605 | 1,267 | 15 % | 1,224 | 14 % | 1,285 | 15 % | 1,242 | 14 % |
| Average | 4,356 | 1,034 | 22 % | 875 | 19 % | 1,041 | 23 % | 875 | 19 % |
The second experiment shows the relationship between the metadata size and the time required to read and parse the DICOM study. In this experiment, the time required to read and load each study stored in the formats described above was measured. For this experiment, the time required to construct the dcm4che2 DICOM objects was recorded. Dcm4che2 was chosen to achieve a fair comparison independent of the normalization process. The results of this experiment are presented in Table 4. Finally, the performance of the normalization algorithm was evaluated. Table 5 contains the time required to build the study model described in the “Materials and Methods” section with and without normalization. The experiments were done using a quad core 2.27-GHz machine with 48 GB of memory and 8 GB allocated heap memory.
Table 4.
Time in milliseconds required for reading DICOM studies stored in different formats. The second column is for SFD format, and the third and fifth are for the MFD and MSD in V1 format. The seventh and ninth columns are for the MFD and MSD in version 2 format. The percentage columns show the read time with respect to the read time of SFD format
| Study name | SFD | MFD (V1) | Percent | MSD (V1) | Percent | MFD (V2) | Percent | MSD (V2) | Percent |
|---|---|---|---|---|---|---|---|---|---|
| SMALLMR | 261.6 | 187 | 71 % | 175 | 67 % | 165.0 | 63 % | 156.9 | 60 % |
| SMALLCT | 361.0 | 466 | 129 % | 460.9 | 128 % | 274.6 | 76 % | 266 | 74 % |
| TESTMR | 615.2 | 403 | 66 % | 377.5 | 61 % | 336.5 | 55 % | 336 | 55 % |
| TESTCT | 985.8 | 1,437 | 146 % | 1,355.7 | 138 % | 798.0 | 81 % | 743.818 | 75 % |
| TESTCTA | 2,069.6 | 2,949 | 142 % | 3,118.8 | 151 % | 1,685.8 | 81 % | 1,391.82 | 67 % |
| BREASTMR | 2,163.8 | 4,109 | 190 % | 4,415 | 204 % | 1,614.4 | 75 % | 1,626.7 | 75 % |
| Average | 1,076 | 1,592 | 124 % | 1,650.48 | 125 % | 812 | 72 % | 753.5 | 68 % |
Table 5.
Performance in milliseconds of the one-pass normalization algorithm. The first column is the time required to construct the study model without normalization. The second is the time required for constructing the study model with normalization using the one-pass normalization algorithm. The last two columns show the speedup in milliseconds and as a percentage
| Study name | Without de-duplication | With de-duplication | Speedup (ms) | Speedup % |
|---|---|---|---|---|
| SMALLMR | 556 | 487 | 69 | 12 % |
| SMALLCT | 858 | 764 | 94 | 11 % |
| TESTMR | 1,270 | 1,199 | 71 | 6 % |
| TESTCT | 2,428 | 2,310 | 118 | 5 % |
| TESTCTA | 5,114 | 4,600 | 514 | 10 % |
| BREASTMR | 5,592 | 5,040 | 552 | 10 % |
| Average | 2,636 | 2,400 | 236 | 9 % |
Discussion
The results in Tables 2 and 3 show a significant reduction in the number of data elements and size of metadata for the MFD and MSD formats compared to the single-frame DICOM. The average number of data elements and average size of the metadata of the MFD v1 formats went down to 18 and 22 % of its original values in the SFD format. The average ratio of the number of data elements and metadata size between the MSD v1 format and the SFD v1 format is 15 and 19 %, respectively. There is no significant difference between the two versions of MSD and MFD formats in terms of the number of data elements and metadata size. The size of a study in a MFD or MSD format with one pixel data element per series is less than its size in the corresponding format with a pixel data element per instance. The reason for this is clear. Increasing the number of pixel data elements requires more space to store the repeated tag number of the pixel data element and consequently increases the metadata size. The difference is equal to the tag size * (number of frames per study - number of series per study).
This size reduction results in a reduction in the time required for reading the DICOM studies. Table 4 shows that the time required to read the studies in MFD and MSD in V2 format is on average about 72 and 68 % of the SFD format, respectively. However, the time required for reading MFD or MSD in V1 format is on average 124 and 125 % of the SFD format, respectively. The study read time depends on two factors. The first is the size of the metadata, and the second is how the pixel data are arranged within the MSD and MFD objects. The first factor decreased the time required to read the DICOM studies in MFD and MSD V2 formats because MFD and MSD have fewer data elements than the SFD format. On the other hand, for the MSD and MFD in V1 formats, the pixel data arrangement was the dominant factor. Concatenating the pixel data for all frames in a series in one data element increased the read time especially for the series that has a large number of frames.
Table 5 shows that the de-duplication algorithm not only does not increase the time required to read and parse the study, it actually reduces the time to read and construct the study object by about 10 % less than the time required to create it without de-duplication. This is because all attributes are added to the non-normalized study model, even if they are repeated, which impacts performance because it increases the model size in memory and consequently the time required to construct the non-normalized study object.
Conclusion
Using sequence data elements, it is possible to store a DICOM study in a single MSD object. The MSD format reduces the number of data elements as well as the size of the metadata compared to SFD and MFD. Reducing the metadata size also reduces the time required to read and parse the study. However, the dominant factor that determines the time required to read the study is the method used to arrange the pixel data within the MFD and MSD objects. Concatenating the pixel data for all frames within a series in a single-pixel data element increases dramatically the time required to read the study. On the other hand, storing the pixel data of each frame in a separate pixel data element within the MFD or MSD object does not create a performance issue. The one-pass de-duplication algorithm is able to significantly reduce the size of the study metadata. In addition, the algorithm has no overhead due to its efficient design and implementation.
Footnotes
Note: DICOM Work Group 6 currently has an ad hoc committee that is trying to create relaxed multi-frame objects that will allow older CT, MR, and PET studies to be converted to series-level objects that reduce duplication and improve transmission time.
Contributor Information
Mahmoud Ismail, Phone: 443-453-6489, Email: maismail@cs.jhu.edu.
James Philbin, Email: james.philbin@jhmi.edu.
References
- 1.DICOM: DICOM standard. DICOM (Digital Imaging and Communications in Medicine), Part 3: Information Object Definitions, Rosslyn, VA 2011. http://medical.nema.org/standard.html. Accessed June 2012