Table 1.
Significant SNPs obtained according to 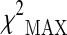 test in the stage 2 sample
test in the stage 2 sample
| SNP ID | dbSNP RS | Variation | Position | Gene | Best model | 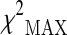 |
p value best model | p valuea |
|---|---|---|---|---|---|---|---|---|
| 6 | rs282070 | G/C | Intron 1 | MAP3K7 | Dominant | 13,939 | 5 × 10−4 | 0.0011 |
| 145 | rs2111699 | A/G | Intron 1 | GSTZ1 | Recessive | 12,511 | 3 × 10−4 | 0.0014 |
| 306 | rs11066301 | A/G | Intron 1 | SHP2 | Dominant | 8,444 | 0.005 | 0.0111 |
| 189 | rs1042571 | C/T | 3′ UTR | POMC | Recessive | 4,413 | 0.046 | 0.0874 |
| 103 | rs3212948 | C/G | Intron 3 | ERCC1 | Recessive | 4,024 | 0.048 | 0.1062 |
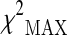 represents the maximum of the three chi-square values
represents the maximum of the three chi-square values
ap value was adjusted for multiple testing of different genetic models obtained by the proposed permutational approach
