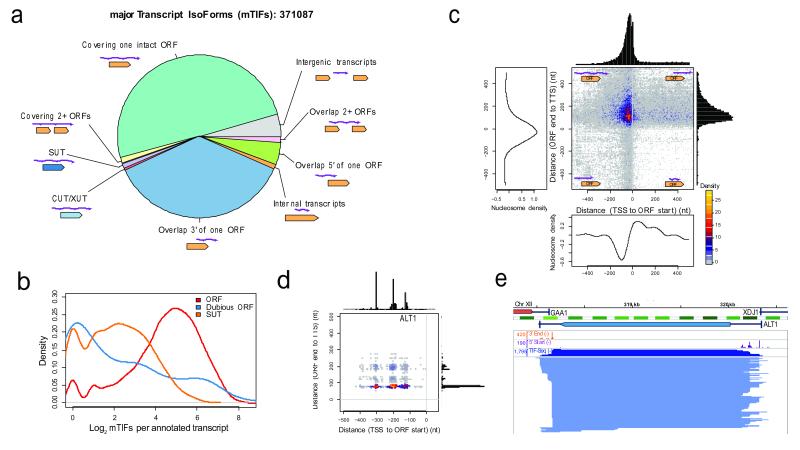
Figure 2. Extensive isoform diversity revealed among overlapping RNA populations, both at the genomic and single-gene level.
a, Categories of mTIFs identified in glucose and galactose. XUT, XRN1-sensitive unstable transcript. b, Log2-scale distribution of clustered mTIFs per annotated transcript that cover characterized or uncharacterized ORFs (ORFs), dubious ORFs, or overlap more than 80% of stable unannotated transcripts (SUTs)5. c, Transcript end distance to ORF stop codon (y-axis) vs. transcript start distance to ORF start codon (x-axis) genome-wide, revealing that most mTIFs cover the entire ORF. Decreased nucleosome density28 coincides with peaks in transcript start and end site distributions. d, Boundaries of TIFs covering ALT1 relative to ORF boundaries (as in c). e, Structure of TIFs overlapping ALT1 in glucose. 5′ start, 3′ end, and TIF-Seq coverage in natural scale. Nucleosome and genome annotations as in Fig. 1d.