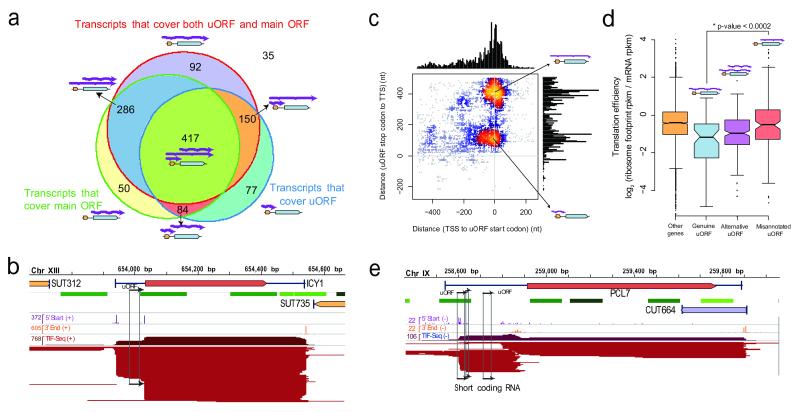
Figure 3. Transcript isoforms with varying regulatory elements and independent short coding RNAs.
a, Number of genes whose mTIFs overlap with previously annotated upstream ORFs (uORFs) and their associated (main) ORFs15. b, ICY1 transcripts in glucose display alternative presence of uORFs (marked with arrows). c, Genome-wide plot of uORF-containing mTIFs: transcript end distance to uORF stop codon (y-axis) vs. transcript start distance to uORF start codon (x-axis). Small coding RNAs previously misannotated as uORFs represent a separate population of short overlapping RNAs. d, Genes with mTIFs that always contain uORFs display lower translation efficiency16 than those for which the uORF is independently transcribed. Genes with alternative presence of uORFs (e.g., ICY1) display intermediate translation efficiency. Significance was computed using the Wilcoxon rank sum test with continuity correction. e, Example of an scRNA that was previously misannotated as a uORF in the PCL7 locus (glucose data shown).