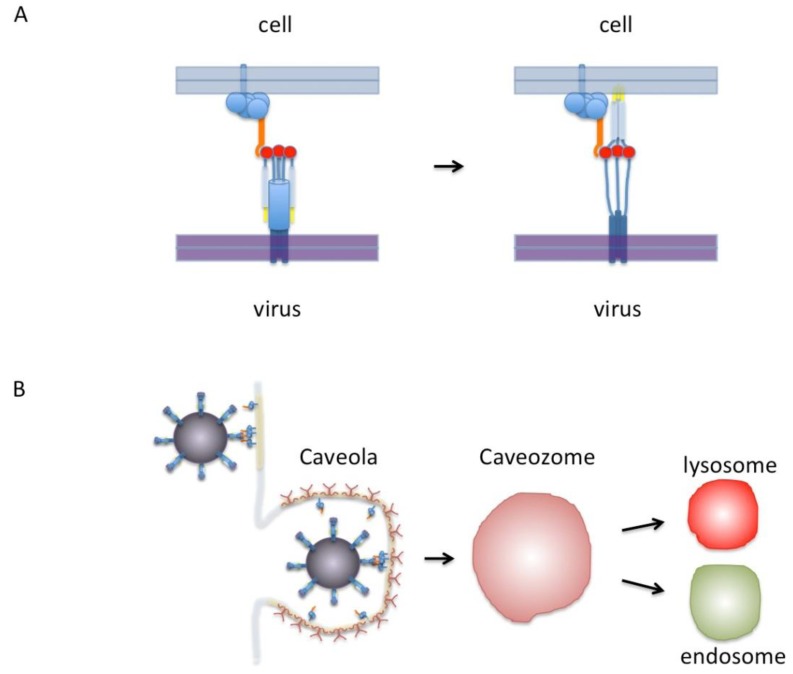
Figure 1.
(A) Model of RSV F-protein binding nucleolin. On the left, the F-protein is shown in its trimeric pre-fusion conformation. The red circles are putative nucleolin binding sites. Nucleolin is shown in orange as part of a protein complex that includes proteins anchored to the membrane by either a transmembrane domain or a GPI anchor. Only one nucleolin molecule is shown binding the F-protein trimer for clarity but in this model as many as three could bind at once. On the right the F-protein is shown in the “extended” conformation with fusion peptides (yellow) inserted into the cell membrane. After this step virus-cell membrane fusion would proceed without nucleolin. (B) Diagram of virus binding to the cell surface. Indicated in light yellow are lipid-rich domains/rafts. The virus is shown in a dark magenta covered with F-protein binding to nucleolin that is preferentially located in lipid-rich rafts or caveolae. A caveola is shown covered with caveolin (dark red). This in turn can enter the cell to form a caveozome and join the endosomal or lysosomal pathway. Our proposed model leaves open the possibilities that viral fusion may occur at the cell surface or in a caveozome/endosome/lysosome. Another possibility (not shown) is that virus enters via clathirin-coated pits (see text).