FIG 3 .
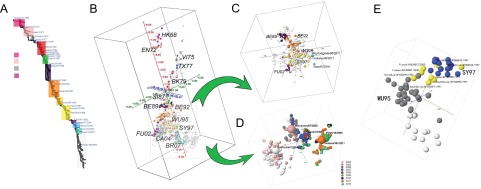
HA1 sequence-based H3N2 antigenic mapping. (A) Phylogenetic tree of H3N2 viruses (1968 to 2012). The selected vaccine strains recommended by the WHO are annotated in this tree. (B) The antigenic map of H3N2 influenza A virus based on nonredundant HA1 sequences (n = 3332). The scoring function was trained by using HI datasets of viruses from 1968 to 2007. The sequences with HI values are marked in color, and others are gray. (C) An antigenic submap of H3N2 viruses isolated from 1991 to 2001. The recently emerged swine origin H3N2v isolates (51) are marked in cyan. (D) An antigenic submap of H3N2 viruses isolated from 2004 to 2012. The viruses are color coded by year, with vaccine strains annotated in enlarged spheres. (E) The map of cluster WU95 and SY97, showing the gradual change. The viruses marked in yellow have 1 or more of the predominant mutations that drive antigenic drift from WU95 to SY97.
