Abstract
Purpose of review
In this review, we summarize the basic principles governing rare variant interpretation in the heritable cardiac arrhythmia syndromes, focusing on recent advances that have led to disease-specific approaches to the interpretation of positive genetic testing results.
Recent findings
Elucidation of the genetic substrates underlying heritable cardiac arrhythmia syndromes has unearthed new arrhythmogenic mechanisms and given rise to a number of clinically meaningful genotype–phenotype correlations. As such, genetic testing for these disorders now carries important diagnostic, prognostic, and therapeutic implications. Recent large-scale systematic studies designed to explore the background genetic ‘noise’ rate associated with these genetic tests have provided important insights and enhanced how positive genetic testing results are interpreted for these potentially lethal, yet highly treatable, cardiovascular disorders.
Summary
Clinically available genetic tests for heritable cardiac arrhythmia syndromes allow the identification of potentially at-risk family members and contribute to the risk-stratification and selection of therapeutic interventions in affected individuals. The systematic evaluation of the ‘signal-to-noise’ ratio associated with these genetic tests has proven critical and essential to assessing the probability that a given variant represents a rare pathogenic mutation or an equally rare, yet innocuous, genetic bystander.
Keywords: Brugada syndrome, genetics, ion channels, long QT syndrome, sudden death
INTRODUCTION
Over the past decade, the discovery that mutations in the genes encoding key cardiac ion channel α-subunit and β-subunit as well as intracellular calcium-handling proteins serve as the primary genetic substrate for a spectrum of heritable cardiac arrhythmia syndromes or ‘cardiac channelopathies’, including long QT syndrome (LQTS), catecholaminergic polymorphic ventricular tachycardia (CPVT), and Brugada syndrome (BrS) [1–3], has broadened our mechanistic understanding of these sudden cardiac death (SCD)-predisposing genetic disorders. Furthermore, these discoveries have given rise to numerous clinically relevant genotype–phenotype correlations, spurring a number of professional societies to recommend the judicious use of clinical genetic testing for the purpose of identifying genetically predisposed individuals with concealed clinical phenotypes and guiding the genotype-specific risk-stratification and clinical management of individuals with clinically definitive disease [4,5,6■■,7■■].
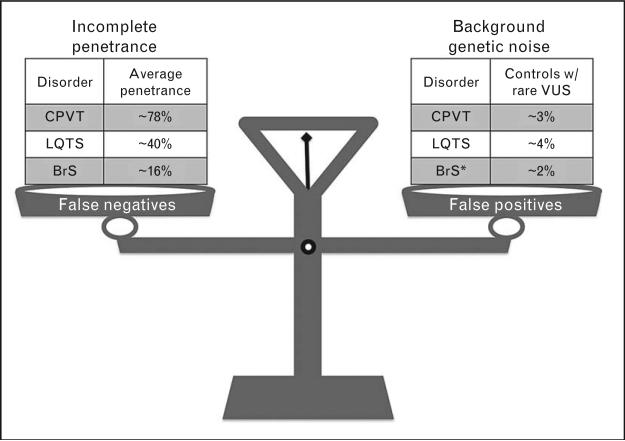
However, as the availability and clinical use of genetic testing increases, so does the probability that rare ‘variants of uncertain significance’ (VUS), alterations in the normal sequence of a gene whose association with disease risk is unknown, will be identified in putative disease-susceptibility genes. As we enter an era of next-generation sequencing, the interpretation of genetic testing results, particularly when the clinical evidence for disease is insufficient or inconclusive, is bound to present an increasingly daunting challenge [8,9]. As with any clinical test, the proper interpretation of genetic testing results requires the careful consideration of all potential sources of both false-positive (e.g., background genetic noise or the frequency of genetic variations in a particular gene in a healthy population) and false-negative (e.g., high prevalence of concealed phenotypes due to incomplete penetrance) results (Fig. 1). Understanding the ‘signal-to-noise’ ratio associated with a given genetic test is particularly important in SCD-predisposing conditions in which the balancing act of distinguishing rare pathogenic mutations from equally rare, yet innocuous, genetic variants can have life-altering implications given that highly effective therapeutic interventions are available, but not entirely devoid of complications or comorbidities, particularly for invasive approaches.
FIGURE 1.
The balancing act involved in the interpretation of heritable cardiac arrhythmia syndrome genetic testing results. Careful consideration of potential sources of false positives (e.g., background genetic noise) and false negatives (e.g., prevalence of concealed phenotypes due to incomplete penetrance) along with the patient's entire clinical picture is necessary to effectively interpret reportedly positive long QT syndrome (LQTS), catecholaminergic polymorphic ventricular tachycardia (CPVT), and Brugada syndrome (BrS) genetic testing results. * indicates that the BrS background noise rate of 2% is just with respect to SCN5A-associated BrS1 in whites. The background noise rate would be much higher when considering BrS1-11 in total and in minorities.
In this review, we describe the basic principles governing rare variant interpretation, present evidence-based algorithms designed to aid in the interpretation of genetic testing results, and lastly detail the clinical utility of a properly interpreted positive genetic test in the diagnosis and clinical management of individuals with these potentially lethal, yet highly treatable, genetic disorders.
General principles of rare variant interpretation
Due to the presence of variable/incomplete disease penetrance and the existence of background genetic noise (Fig. 1), the interpretation of genetic testing should, with the exception of certain well-established disease-causative mutations, always be viewed as probabilistic, rather than deterministic/binary, in nature. When interpreted in the context of the overall clinical picture by experienced personnel (e.g., molecular/genetic cardiologists, clinical/cardiovascular geneticists, or genetic counselors), there are several criteria that can be used to support labeling a given variant as a ‘potentially disease-causative mutation’. These criteria often include, but are not necessarily limited to, cosegregation of the variant with disease; absence or extreme rarity of the variant in control cohorts or publicly available exome/genome datasets; nonsense, frameshift, or insertion/deletion mutations that lead to truncated protein products; localization of nonsynonymous single nucleotide variants (nsSNVs) to highly conserved amino acid residues/key functional domains and/or result in radical amino acid substitutions (i.e., phylogenetic/physicochemical properties); and evidence of perturbed ion channel function through in-vitro functional studies [10,11]. Although these criteria are quite helpful, it is important to remember that satisfying any one of these particular criteria does not necessarily equate to a designation as a definite, pathogenic mutation. In fact, in-silico tools like Sorting Intolerant from Tolerant (SIFT), PolyPhen2, and so on, that assess a variant's likelihood of pathogenicity based on phylogenetic and/or physicochemical properties, must be used with great caution because in isolation their false-positive rate can be quite high, especially when applied to LQTS genetic testing [12].
When these general principles fail to provide sufficient cumulative evidence to label a given variant as ‘pathogenic/disease-causative’, the variant should be deemed a VUS until additional information (e.g., disease-specific considerations, functional studies, etc.) to either confirm or deny its pathogenicity can be obtained. Importantly, the presence of a VUS should never be used to establish a diagnosis in the index case or for confirmatory genetic testing of potentially at-risk relatives.
Long QT syndrome
Congenital LQTS is a genetically heterogeneous disorder of myocardial repolarization with an estimated prevalence as high as one in 2000 persons [13]. Clinically, LQTS is objectively characterized by a prolonged heart rate-corrected QT interval (QTc) that exceeds the 99th percentile values for otherwise healthy men (> 470 ms) and women (> 480 ms) on 12-lead ECG and an increased risk of syncope, seizures, and sudden death secondary to torsades de pointes, the characteristic form of polymorphic ventricular fibrillation observed in LQTS [14]. However, the clinical diagnosis of LQTS is often clouded by the genetic phenomena of incomplete penetrance and variable expressivity [15–17], as evidenced by the 10–40% of genotype-positive individuals who fail to display overt QT prolongation on resting ECG [15,18].
At present, mutations in 13 distinct LQTS-susceptibility genes have been implicated in the pathogenesis of the disorder (Table 1 [19–47]). However, 60–75% of clinically definitive LQTS patients harbor a mutation in one of three major LQTS-susceptibility genes: the KCNQ1-encoded Kv7.1 potassium channel (LQT1, 30–35%), the KCNH2-encoded Kv11.1/hERG potassium channel (LQT2, 25–30%), or the SCN5A-encoded Nav1.5 sodium channel (LQT3, 5–10%) [24,48]. Inclusion of the remaining 10 minor LQTS-susceptibility genes increases the genetic yield by less than 5%, but also increases the likelihood of encountering a false-positive. Additionally, reflex testing for copy number variations/larger genomic rearrangements in KCNQ1 and KCNH2 is available and may increase the overall yield to approximately 80–85% [49,50]. At minimum, all clinically available genetic tests include the three major LQTS-susceptibility genes, and the expected yield of targeted (LQT1–3) and comprehensive (LQT1–LQT13) genetic testing in index cases with unequivocal evidence of disease is summarized in Table 2 [7■■,18].
Table 1.
Genetic basis of cardiac channelopathies
| Gene (genotype) | Locus | Protein | Frequency | Ref. |
|---|---|---|---|---|
| LQTS | ||||
| KCNQ1 (LQT1)a | 11p15.5 | Kv7.1 | 30–35% | [19] |
| KCNH2 (LQT2)a | 7q35-46 | Kv11.1 | 25–30% | [20] |
| SCN5A (LQT3)a | 3p21-p24 | Nav1.5 | 5–10% | [21] |
| ANKB (LQT4)a | 4q25-q27 | Ankyrin B | Rare | [22] |
| KCNE1 (LQT5)a | 21q22.1 | MinK | Rare | [23] |
| KCNE2 (LQT6)a | 21q22.1 | MiRP1 | Rare | [24] |
| KCNJ2 (LQT7/ATS1)a | 17q23 | Kir2.1 | Rare | [25] |
| CACNA1C (LQT8/TS1)a | 12p13.3 | Cav1.2 | Rare | [26] |
| CAV3 (LQT9)a | 3p25 | Caveolin 3 | Rare | [27] |
| SCN4B (LQT10)a | 11q23.3 | Nav1.5 β4-subunit | Rare | [28] |
| AKKAP9 (LQT11)a | 7q21-q22 | Yotiao | Rare | [29] |
| SNTA1 (LQT12)a | 20q11.2 | Syntrophin-α1 | Rare | [30] |
| KCNJ5 (LQT13) | 11q24 | Kir3.4 | Rare | [31] |
| CPVT | ||||
| RYR2 (CPVT1)a | 1q42.1-43 | Ryanodine receptor 2 | 50–60% | [32] |
| CASQ2 (CPVT2)a | 1p13.3-p11 | Calsequestrin 2 | 1–2% | [33] |
| KCNJ2 (CPVT3)a | 17q23 | Kir2.1 | 10% | [34] |
| BrS | ||||
| SCN5A (BrS1)a | 3p21-p24 | Nav1.5 | 20–30% | [35] |
| GPD1L (BrS2)a | 3q22.3 | Glycerol-3-phosphate dehydrogenase 1-like | Rare | [36] |
| CACNA1C (BrS3)a | 12p13.3 | Cav1.2 | 6.6% | [37] |
| CACNB2 (BrS4)a | 10p12 | Cav1.2 β2-subunit | Rare | [38] |
| SCN1B (BrS5)a | 19q13.1 | Nav1.5 β1-subunit | Rare | [39] |
| KCNE3 (BrS6)a | 11q13.4 | MiRP2 | Rare | [40] |
| SCN3B (BrS7)a | 11q24.1 | Nav1.5 β3-subunit | Rare | [41] |
| KCNJ8 (BrS8)a | 12p12.1 | Kir6.1 | 2% | [42,43] |
| CACNA2D1 (BrS9) | 7q21-q22 | Cav1.2 α2/δ1-subunit | Rare | [44] |
| KCND3 (BrS10)a | 1p13.2 | Kv4.3 | Rare | [45,46] |
| MOG1 (BrS11) | 17p13.1 | Mog1 | Rare | [47] |
Rare is defined as contributing to fewer than 1% of cases. Although tentative disease subtypes are provided in parenthesis, we strongly recommend annotating only common disease subtypes numerically. For rare genetic subtypes, we suggest that a descriptor such as SNTA1-LQTS instead of LQT12 is preferable, as no consensus exists on the numerical nomenclature for many minor subtypes. ATS, Andersen–Tawil syndrome; BrS, Brugada syndrome; CPVT, catecholaminergic polymorphic ventricular tachycardia; LQTS, long QT syndrome; TS, Timothy syndrome.
Commercial genetic testing for these genes is available from at least one company.
Table 2.
Clinical impact and considerations for genetic testing in cardiac channelopathy index cases
| Epidemiologic considerations |
Clinical impact |
Practical considerations |
|||||
|---|---|---|---|---|---|---|---|
| Disorder | Prevalence | Annual event rate | Diagnostic | Prognostic | Therapeutic | Yield of genetic testa | Signal-to-noise ratiob |
| LQTS | 1 : 2000 | 0.3–0.6% | Strong | Strong | Moderate | 75% (80%) | 19 : 1 |
| CPVT | 1 : 7000 | 3.1% | Strong | Weak | Negligible | 60% (70%) | 20 : 1 |
| BrS | 5 : 10000 | 1.4% | Moderate | Weak | Negligible | 20% (30%) | 10 : 1 |
BrS, Brugada syndrome; CPVT, catecholaminergic ventricular tachycardia; LQTS, long QT syndrome.
Yield of genetic test represents published/unpublished estimates derived from unrelated cases with robust clinical phenotypes. The first number represents the yield of the targeted major susceptibility-gene screen for whites, whereas numbers in parentheses represent the total yield of all known disease-susceptibility genes included in commercially available panels for whites. There is currently insufficient evidence to derive genetic yields for minority populations.
The signal-to-noise ratio is calculated by dividing the yield in cases by the background rate of variants of uncertain significance (VUS) in controls.
The clinical impact and practical consideration sections are reproduced in part from [7■■].
Recent studies have demonstrated that approximately 4% of ostensibly healthy white individuals and 6–8% of black individuals harbor a rare, amino acid-altering genetic variant in one of the three major LQTS-susceptibility genes (KCNQ1, KCNH2, or SCN5A) [51]. Coupling the established rate of innocuous background genetic variation with large compendia of rare variants identified in clinically definite LQTS cases has allowed the calculation of a so-called signal-to-noise ratio associated with targeted genetic testing (Table 2) and yielded a number of observations that have improved how the presence of a VUS in a major LQTS-susceptibility gene should be interpreted.
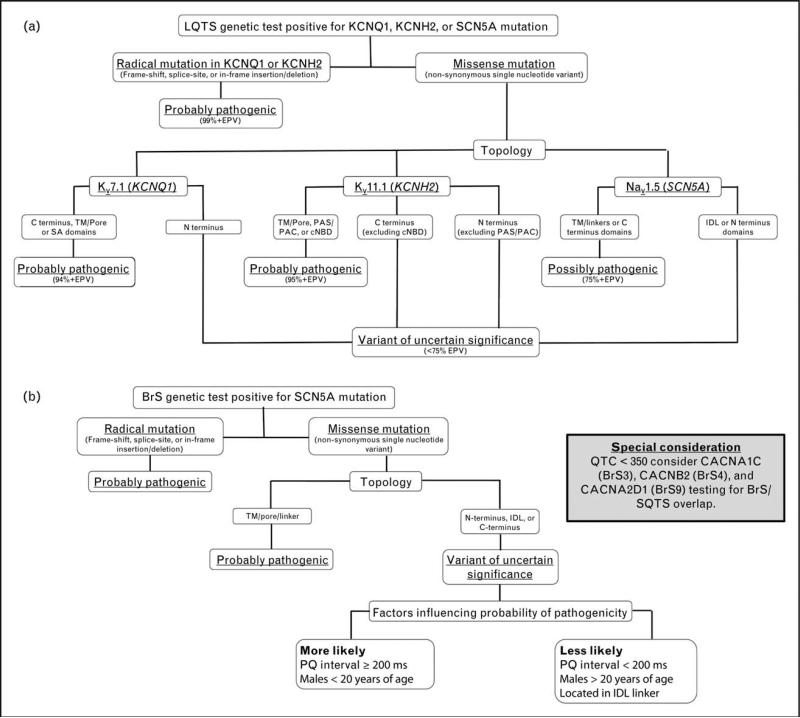
Specifically, certain mutation types (e.g., radical/truncating) and missense mutation localizing to specific topological structure-function domains of the Kv11.1/hERG, Kv7.1, and Nav1.5 channels (e.g., pore/transmembrane regions) carry a high (>95%) estimated predictive value (EPV), suggesting that, when identified in a case with high clinical probability for the contemplated clinical diagnosis, these mutations carry a high genetic probability of being disease-causative [51]. Collectively, this body of evidence has been used to synthesize a topology-driven algorithm designed to aid in the probabilistic interpretation of positive LQTS genetic testing results (Fig. 2a).
FIGURE 2.
Evidence-based algorithms designed to aid in the interpretation of positive long QT syndrome and Brugada syndrome genetic testing results. (a) Algorithm for interpreting a positive long QT syndrome (LQTS) genetic test. Radical mutations that significantly alter/truncate Kv7.1 or Kv11.1, such as insertions/deletions, alteration of intronic/exonic splice site boundaries, and nonsense mutations, are probably LQTS-associated. Those rare, absent in controls, nonsynonymous single nucleotide variants (nsSNVs; missense mutations) that localize to Kv7.1 (TM/pore, SA, or C terminal domains), Kv11.1 (TM/Pore, PAS/PAC, or cNBD), or Nav1.5 (TM/pore/linker or C terminus) are probably or possibly pathogenic. Variants outside these topological structure-function domains are deemed to be variants of uncertain significance (VUS), unless additional evidence is present (e.g., cosegregation with disease, LQTS-like electrophysiological phenotype, etc.). cNBD, cyclic nucleotide binding domain; EPV, estimated predictive value; IDL, interdomain linker; PAC, per-arnt-sim C-terminal associated; PAS, per-arnt-sim; SA, subunit assembly; TM, transmembrane. (b) Algorithm for interpreting a positive Brugada syndrome (BrS) genetic test. Radical mutations that significantly alter/truncate Nav1.5 protein structure, such as insertions/deletions, alteration of intronic/exonic splice site boundaries, and nonsense mutations, are probably pathogenic. Those rare, absent in controls, nsSNVs (missense mutations) that localize to the Nav1.5 TM/pore/linker are probably pathogenic. Variants outside this topological structure-function domain, particularly those residing in the first interdomain linker (IDL), are of uncertain disease relevance. When such a VUS is identified in an individual with a PQ interval at least 200 ms or a male less than 20 years of age, its probability of pathogenicity increases but not sufficiently so to be declared a BrS1-associated mutation without additional evidence. IDL, interdomain linker; TM, transmembrane domain.
Based on current Heart Rhythm Society (HRS)/European Heart Rhythm Association (EHRA) guidelines, comprehensive or targeted LQTS genetic testing is recommended for any individual with a strong clinical suspicion of LQTS based on clinical/family history and electrocardiographic phenotype or an asymptomatic individual with unexplained QT prolongation (> 480 before puberty and > 500 after puberty) [7■■]. Mutation-specific testing/cascade screening is recommended for the relatives of genotype-positive LQTS index patients, even for family members with negative clinical/electrocardiographic phenotypes [7■■]. Although genetic testing is not needed to establish a diagnosis of LQTS in the setting of a robust clinical phenotype, genotype is one of many factors to consider when assessing the risk of SCD and selecting appropriate therapeutic interventions [52]. Specifically, genotype has proven particularly useful in predicting the efficacy of β-blocker pharmacotherapy as β-blockade has been shown to be extremely protective in LQT1 patients and moderately protective in LQT2 patients [53]. In contrast, targeting the pathogenic late sodium current with a combination of a β-blocker (propranolol) and possibly mexiletine, flecainide, or ranolazine represents the preferred pharmacotherapy option for LQT3 patients [54,55]. Nevertheless, it should be stressed that all risk-stratification/management decisions, particularly prophylactic implantable cardioverter defibrillator (ICD) implantation in LQT3 patients, require the careful consideration of the patient's entire clinical picture, which includes both genetic and nongenetic risk factors such as 15 years of age or less, female sex, and QTc more than 500 ms.
Catecholaminergic ventricular tachycardia
CPVT is a genetic disease of intracellular calcium handling that affects an estimated one in 7000 to one in 10 000 individuals and is associated with an increased risk of syncope and SCD secondary to adrenergically mediated ventricular arrhythmias [56–58]. In general, CPVT is associated with a normal resting ECG and diagnosed on the basis of clinical history and the induction of significant ventricular ectopy, including the diagnostic hallmark of bidirectional or polymorphic ventricular tachycardia, during exercise or catecholamine stress testing [58,59]. In contrast to other cardiac channelopathies, the penetrance and expressivity of CPVT appears to be much higher, with an overall disease penetrance reported to be approximately 80% [60,61■] and a positive family history of SCD present in up to 60% of families harboring mutations in RYR2 [56].
At present, mutations in three distinct CPVT-susceptibility genes have been associated with the pathogenesis of CPVT (Table 1). Approximately 60–65% of CPVT index cases harbor a mutation in the RYR2-encoded ryanodine receptor 2/intracellular calcium release channel (CPVT1), whereas mutations in the minor genes (CASQ2/CPVT2 and KCNJ2/CPVT3) are identified in fewer than 5% of cases [56,57,62]. The overall yield and signal-to-noise ratio of CPVT genetic testing are summarized in Table 2.
At present, over 100 CPVT1-causative mutations have been reported and these mutations tend to localize to three specific clusters/regions of the RyR2 protein. The fact that nearly all of the known CPVT1 mutations localize to only 45 exons of the massive and hypomorphic 105-exon-containing RYR2 gene has led several groups to propose targeted/tiered screening approaches [57]. However, the identification of CPVT1-causative mutations that reside outside these ‘hotspot’ exons has led to some concern that targeted/tiered screening may be suboptimal [34]. As such, no consensus or clear definition of the most appropriate strategy for RYR2 screening currently exists.
Given the relative paucity of background genetic variation in RYR2, particularly within hotspot exons, no additional CPVT-specific criteria for the interpretation of a positive genetic test currently exist. Based on current HRS/EHRA guidelines, CPVT genetic testing is recommended for any individual with a strong clinical suspicion of CPVT based on history and electrocardiographic phenotype observed during exercise/catecholamine provocation testing for the primary purpose of identifying the underlying CPVT-causative mutation needed to facilitate the recommended mutation-specific screening of all potentially at-risk first-degree and second-degree relatives, including newborns/infants given the known association between CPVT and sudden infant death syndrome [7■■]. Genotype presently has no bearing on the risk-stratification or selection of management strategies in patients with CPVT.
Brugada syndrome
BrS is a rare heritable cardiac arrhythmia syndrome with an estimated prevalence of five in 10 000 individuals of European descent, but may be more prevalent in those of Asian descent [63]. Clinically, BrS is characterized by the presence of coved-type ST-segment elevation and inverted T-waves (type 1 BrS ECG pattern) in the right precordial leads (V1–V3) and an increased risk of syncope and SCD secondary to re-entrant ventricular tachycardia/ventricular fibrillation [64,65]. BrS predominantly affects post-pubertal men between 20 and 50 years of age with SCD generally occurring during sleep [66]. Due to the transient and dynamic nature of BrS ECG pattern, sodium channel blockers (e.g., flecainide, procainamide, and ajmaline) may precipitate a type 1 BrS ECG pattern in order to clarify the diagnosis in patients with suspected disease [63]. In comparison to other cardiac channelopathies, overall disease penetrance in BrS appears to be quite low, as evidenced by the observation that on average only 16% (range 12.5–50%) of SCN5A mutation-positive individuals across 52 BrS families featured spontaneous/sodium channel blocked-induced ST-segment elevation and/or a history of cardiac events [67].
Over the past 20 years, mutations in at least 11 distinct BrS-susceptibility genes have been identified (Table 1). Loss-of-function mutations in the SCN5A-encoded Nav1.5 sodium channel represent the most common genetic substrate (BrS1), accounting for approximately 15–30% of all BrS cases [67–69]. An additional 10–15% of BrS cases, particularly individuals with ST-segment elevation and a shortened QT interval (< 330 ms for men and < 340 ms for women), may be linked to perturbation of the L-type calcium channel (Table 1) [37,44]. However, recent evidence suggests that, in the absence of concomitant ST-segment elevation and short QT interval, a BrS2 through BrS11 genotype is collectively observed in fewer than 5% of cases [70■]. However, it should be noted that this observation might also be cohort-specific.
Due to the combined rarity of the minor BrS genotypes, only BrS1 (SCN5A) targeted BrS genetic testing has been deemed to be clinically useful at the present time [6■■,7■■] and the expected genetic yield/signal-to-noise ratio associated with targeted testing is summarized in Table 2. Although no studies that specifically address the issue of SCN5A variant interpretation in BrS exist, evidence derived from the study of a large international compendium of BrS-associated SCN5A mutations [71] and the systematic assessment of mutations in the 12 BrS-susceptibility genes in a large unrelated cohort of BrS patients [70■] have yielded several important observations/genotype–phenotype correlations that may aid in the interpretation of BrS genetic testing results. An algorithm synthesized from this body of evidence is outlined in Fig. 2b.
Based on current HRS/EHRA guidelines, genetic testing for SCN5A alone is useful for most individuals with a strong clinical suspicion of BrS based on clinical/family history and or electrocardiographic phenotype with the primary goal of identifying a BrS-causative mutation in the index case that would facilitate the recommended mutation-specific screening of appropriate relatives [6■■,7■■,70■]. Phenotype-guided genetic analysis of CACNA1C, CACNB2, and CACNA2D1 may be useful in BrS patients who also feature a short QT interval [70■]. However, like CPVT, no clear prognostic or therapeutic implications are associated with positive genetic testing results.
Other heritable cardiac arrhythmia syndromes
Genetic testing for cardiac channelopathies such as short QT syndrome (SQTS), familial atrial fibrillation (FAF), and cardiac conduction disease (CCD) less frequently encountered in clinical practice is available. However, the lower contribution of known disease-susceptibility genes to the pathogenesis of these disorders currently limits the clinical utility of these tests. As such, few insights exist to guide the interpretation of SQTS, FAF, or CCD genetic testing results.
CONCLUSION
Given that clinical genetic testing should be viewed as probabilistic rather than deterministic in nature, the accurate interpretation of genetic testing results requires careful consideration of both genetic and clinical factors. The elucidation of the spectrum of healthy genetic variation inherent in channelopathy-susceptibility genes that comprises the background genetic noise associated with clinical genetic testing for these disorders has provided important insights that now guide the interpretation of positive genetic test results. With the recent completion of large-scale exome/genome sequencing projects (e.g., 1000 Genomes Project), we are beginning to realize that the prevalence of functionally relevant genetic variation within putative channelopathy-susceptibility genes may be higher than previously anticipated. Cautious scrutiny of this evolving compendium of human genetic variation in health coupled with the use of next-generation sequencing technology to thoroughly explore the genetic architecture underlying the heritable cardiac arrhythmia syndromes promises to provide clinically meaningful insights that will improve the interpretation of positive genetic testing results in the future for these potentially lethal, yet highly treatable, genetic disorders.
KEY POINTS.
Clinical genetic testing for heritable cardiac arrhythmia syndromes must be viewed as probabilistic, rather than deterministic, and always interpreted in the context of a patient's entire clinical picture.
Rare variant interpretation is a balancing act that requires careful consideration of both the prevalence of genotype-positive individuals with concealed clinical phenotypes secondary to incomplete penetrance and variable expressivity and the estimated one in 25 otherwise healthy individuals expected to harbor a rare, and most likely innocuous, background genetic variant within one of the three major channelopathysusceptibility genes.
Radical/truncating mutations and missense mutations residing within critical structure-function domains (e.g., transmembrane/pore) of the cardiac ion channels encoded by the one major BrS-susceptibility and three major LQTS-susceptibility gene(s) are probably pathogenic when identified in patients with a robust clinical phenotype.
The judicious use and proper interpretation of clinical genetic tests offer the potential to identify potentially at-risk relatives, contribute to risk-stratification, and guide the selection of appropriate therapeutic interventions.
Acknowledgements
Research into the genetic causes of channelopathic sudden cardiac death is supported by the National Institutes of Health (R01-HD42569), a Fondation Leducq Award for the ‘Alliance for Calmodulin Kinase Signaling in Heart Disease’, and the Windland Smith Rice Sudden Comprehensive Sudden Cardiac Death Program. J.R.G. is supported by a National Heart, Lung, and Blood Institute Kirchstein NRSA Individual Predoctoral MD/PhD Fellowship (F30-HL106993).
Footnotes
Conflicts of interest
M.J.A. is a consultant for Biotronik, Boston Scientific, Medtronic, St Jude Medical and Transgenomic. Intellectual property derived from M.J.A.'s research program resulted in license agreements in 2004 between Mayo Clinic Health Solutions (formerly Mayo Medical Ventures) and PGxHealth (formerly Genaissance Pharmaceuticals, now recently acquired by Transgenomic). J.R.G. declares no conflict of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
■ of special interest
■■ of outstanding interest
Additional references related to this topic can also be found in the Current World Literature section in this issue (pp. 82–83).
- 1.Ruan Y, Liu N, Priori SG. Sodium channel mutations and arrhythmias. Nat Rev Cardiol. 2009;6:337–348. doi: 10.1038/nrcardio.2009.44. [DOI] [PubMed] [Google Scholar]
- 2.Cerrone M, Priori SG. Genetics of sudden death: focus on inherited channelopathies. Eur Heart J. 2011;32:2109–2118. doi: 10.1093/eurheartj/ehr082. [DOI] [PubMed] [Google Scholar]
- 3.Giudicessi JR, Ackerman MJ. Potassium-channel mutations and cardiac arrhythmias: diagnosis and therapy. Nat Rev Cardiol. 2012;9:319–332. doi: 10.1038/nrcardio.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zipes DP, Camm AJ, Borggrefe M, et al. ACC/AHA/ESC 2006 guidelines for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: a report of the American College of Cardiology/American Heart Association Task Force and the European Society of Cardiology Committee for Practice Guidelines (Writing Committee to Develop Guidelines for Management of Patients With Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death). J Am Coll Cardiol. 2006;48:e247–e346. doi: 10.1016/j.jacc.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Heart Rhythm UK Familial Sudden Death Syndromes Statement Development Group Clinical indications for genetic testing in familial sudden cardiac death syndromes: an HRUK position statement. Heart. 2008;94:502–507. doi: 10.1136/hrt.2007.127761. [DOI] [PubMed] [Google Scholar]
- 6■■.Gollob MH, Blier L, Brugada R, et al. Recommendations for the use of genetic testing in the clinical evaluation of inherited cardiac arrhythmias associated with sudden cardiac death: Canadian Cardiovascular Society/Canadian Heart Rhythm Society joint position paper. Can J Cardiol. 2011;27:232–245. doi: 10.1016/j.cjca.2010.12.078. [The Canadian Cardiology Society/Canadian Heart Rhythm Society guidelines largely echo the consensus of HRS/EHRA; however, there are some differing opinions that readers may want to note. Most germane to the current subject matter is the genetic testing of patients with only a type 1 BrS pattern who do not fulfill the official criteria for BrS.] [DOI] [PubMed] [Google Scholar]
- 7■■.Ackerman MJ, Priori SG, Willems S, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Heart Rhythm. 2011;8:1308–1339. doi: 10.1016/j.hrthm.2011.05.020. [The HRS/EHRA guidelines are the most up-to-date guidelines regarding genetic testing in the cardiac channelopathies written and endorsed by a global panel of heart rhythm specialists.] [DOI] [PubMed] [Google Scholar]
- 8.Evans JP, Skrzynia C, Burke W. The complexities of predictive genetic testing. BMJ. 2001;322:1052–1056. doi: 10.1136/bmj.322.7293.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter DJ, Khoury MJ, Drazen JM. Letting the genome out of the bottle: will we get our wish? N Engl J Med. 2008;358:105–107. doi: 10.1056/NEJMp0708162. [DOI] [PubMed] [Google Scholar]
- 10.Cotton RG, Scriver CR. Proof of ‘disease causing’ mutation. Hum Mutat. 1998;12:1–3. doi: 10.1002/(SICI)1098-1004(1998)12:1<1::AID-HUMU1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 11.Ingles J, Zodgekar PR, Yeates L, et al. Guidelines for genetic testing of inherited cardiac disorders. Heart Lung Circ. 2011;20:681–687. doi: 10.1016/j.hlc.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 12.Giudicessi JR, Kapplinger JD, Tester DJ, et al. Phylogenetic and physico-chemical analyses enhance the classification of rare non-synonymous single nucleotide variants in type 1 and 2 Long QT syndrome. Circ Cardiovasc Genet. 2012;5:519–528. doi: 10.1161/CIRCGENETICS.112.963785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz PJ, Stramba-Badiale M, Crotti L, et al. Prevalence of the congenital long-QT syndrome. Circulation. 2009;120:1761–1767. doi: 10.1161/CIRCULATIONAHA.109.863209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita H, Wu J, Zipes DP. The QT syndromes: long and short. Lancet. 2008;372:750–763. doi: 10.1016/S0140-6736(08)61307-0. [DOI] [PubMed] [Google Scholar]
- 15.Priori SG, Napolitano C, Schwartz PJ. Low penetrance in the long-QT syndrome: clinical impact. Circulation. 1999;99:529–533. doi: 10.1161/01.cir.99.4.529. [DOI] [PubMed] [Google Scholar]
- 16.Vincent GM, Timothy KW, Leppert M, Keating M. The spectrum of symptoms and QT intervals in carriers of the gene for the long-QT syndrome. N Engl J Med. 1992;327:846–852. doi: 10.1056/NEJM199209173271204. [DOI] [PubMed] [Google Scholar]
- 17.Amin AS, Giudicessi JR, Tijsen AJ, et al. Variants in the 3’ untranslated region of the KCNQ1-encoded Kv7.1 potassium channel modify disease severity in patients with type 1 long QT syndrome in an allele-specific manner. Eur Heart J. 2012;33:714–723. doi: 10.1093/eurheartj/ehr473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tester DJ, Will ML, Haglund CM, Ackerman MJ. Effect of clinical phenotype on yield of long QT syndrome genetic testing. J Am Coll Cardiol. 2006;47:764–768. doi: 10.1016/j.jacc.2005.09.056. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Curran ME, Splawski I, et al. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- 20.Curran ME, Splawski I, Timothy KW, et al. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, Shen J, Splawski I, et al. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 22.Mohler PJ, Schott JJ, Gramolini AO, et al. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature. 2003;421:634–639. doi: 10.1038/nature01335. [DOI] [PubMed] [Google Scholar]
- 23.Duggal P, Vesely MR, Wattanasirichaigoon D, et al. Mutation of the gene for IsK associated with both Jervell and Lange-Nielsen and Romano-Ward forms of Long-QT syndrome. Circulation. 1998;97:142–146. doi: 10.1161/01.cir.97.2.142. [DOI] [PubMed] [Google Scholar]
- 24.Splawski I, Shen J, Timothy KW, et al. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102:1178–1185. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 25.Plaster NM, Tawil R, Tristani-Firouzi M, et al. Mutations in Kir2.1 cause the developmental and episodic electrical phenotypes of Andersen's syndrome. Cell. 2001;105:511–519. doi: 10.1016/s0092-8674(01)00342-7. [DOI] [PubMed] [Google Scholar]
- 26.Splawski I, Timothy KW, Sharpe LM, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Vatta M, Ackerman MJ, Ye B, et al. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation. 2006;114:2104–2112. doi: 10.1161/CIRCULATIONAHA.106.635268. [DOI] [PubMed] [Google Scholar]
- 28.Medeiros-Domingo A, Kaku T, Tester DJ, et al. SCN4B-encoded sodium channel beta4 subunit in congenital long-QT syndrome. Circulation. 2007;116:134–142. doi: 10.1161/CIRCULATIONAHA.106.659086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Marquardt ML, Tester DJ, et al. Mutation of an A-kinase-anchoring protein causes long-QT syndrome. Proc Natl Acad Sci U S A. 2007;104:20990–20995. doi: 10.1073/pnas.0710527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ueda K, Valdivia C, Medeiros-Domingo A, et al. Syntrophin mutation associated with long QT syndrome through activation of the nNOS-SCN5A macromolecular complex. Proc Natl Acad Sci U S A. 2008;105:9355–9360. doi: 10.1073/pnas.0801294105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Liang B, Liu J, et al. Identification of a Kir3.4 mutation in congenital long QT syndrome. Am J Hum Genet. 2010;86:872–880. doi: 10.1016/j.ajhg.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Priori SG, Napolitano C, Tiso N, et al. Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation. 2001;103:196–200. doi: 10.1161/01.cir.103.2.196. [DOI] [PubMed] [Google Scholar]
- 33.Lahat H, Pras E, Olender T, et al. A missense mutation in a highly conserved region of CASQ2 is associated with autosomal recessive catecholamine-induced polymorphic ventricular tachycardia in Bedouin families from Israel. Am J Hum Genet. 2001;69:1378–1384. doi: 10.1086/324565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tester DJ, Arya P, Will M, et al. Genotypic heterogeneity and phenotypic mimicry among unrelated patients referred for catecholaminergic polymorphic ventricular tachycardia genetic testing. Heart Rhythm. 2006;3:800–805. doi: 10.1016/j.hrthm.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 35.Chen Q, Kirsch GE, Zhang D, et al. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392:293–296. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 36.London B, Michalec M, Mehdi H, et al. Mutation in glycerol-3-phosphate dehydrogenase 1 like gene (GPD1-L) decreases cardiac Na+ current and causes inherited arrhythmias. Circulation. 2007;116:2260–2268. doi: 10.1161/CIRCULATIONAHA.107.703330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antzelevitch C, Pollevick GD, Cordeiro JM, et al. Loss-of-function mutations in the cardiac calcium channel underlie a new clinical entity characterized by ST-segment elevation, short QT intervals, and sudden cardiac death. Circulation. 2007;115:442–449. doi: 10.1161/CIRCULATIONAHA.106.668392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cordeiro JM, Marieb M, Pfeiffer R, et al. Accelerated inactivation of the L-type calcium current due to a mutation in CACNB2b underlies Brugada syndrome. J Mol Cell Cardiol. 2009;46:695–703. doi: 10.1016/j.yjmcc.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe H, Koopmann TT, Le Scouarnec S, et al. Sodium channel beta1 subunit mutations associated with Brugada syndrome and cardiac conduction disease in humans. J Clin Invest. 2008;118:2260–2268. doi: 10.1172/JCI33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delpon E, Cordeiro JM, Nunez L, et al. Functional effects of KCNE3 mutation and its role in the development of Brugada syndrome. Circ Arrhythm Electrophysiol. 2008;1:209–218. doi: 10.1161/CIRCEP.107.748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu D, Barajas-Martinez H, Burashnikov E, et al. A mutation in the beta 3 subunit of the cardiac sodium channel associated with Brugada ECG phenotype. Circ Cardiovasc Genet. 2009;2:270–278. doi: 10.1161/CIRCGENETICS.108.829192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medeiros-Domingo A, Tan BH, Crotti L, et al. Gain-of-function mutation S422L in the KCNJ8-encoded cardiac K(ATP) channel Kir6.1 as a pathogenic substrate for J-wave syndromes. Heart Rhythm. 2010;7:1466–1471. doi: 10.1016/j.hrthm.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barajas-Martinez H, Hu D, Ferrer T, et al. Molecular genetic and functional association of Brugada and early repolarization syndromes with S422L missense mutation in KCNJ8. Heart Rhythm. 2012;9:548–555. doi: 10.1016/j.hrthm.2011.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burashnikov E, Pfeiffer R, Barajas-Martinez H, et al. Mutations in the cardiac L-type calcium channel associated with inherited J-wave syndromes and sudden cardiac death. Heart Rhythm. 2010;7:1872–1882. doi: 10.1016/j.hrthm.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giudicessi JR, Ye D, Tester DJ, et al. Transient outward current (I(to)) gain-of-function mutations in the KCND3-encoded Kv4.3 potassium channel and Brugada syndrome. Heart Rhythm. 2011;8:1024–1032. doi: 10.1016/j.hrthm.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giudicessi JR, Ye D, Kritzberger CJ, et al. Novel mutations in the KCND3-encoded Kv4.3 K+ channel associated with autopsy-negative sudden unexplained death. Hum Mutat. 2012;33:989–997. doi: 10.1002/humu.22058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kattygnarath D, Maugenre S, Neyroud N, et al. MOG1: a new susceptibility gene for Brugada syndrome. Circ Cardiovasc Genet. 2011;4:261–268. doi: 10.1161/CIRCGENETICS.110.959130. [DOI] [PubMed] [Google Scholar]
- 48.Tester DJ, Will ML, Haglund CM, Ackerman MJ. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2005;2:507–517. doi: 10.1016/j.hrthm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 49.Eddy CA, MacCormick JM, Chung SK, et al. Identification of large gene deletions and duplications in KCNQ1 and KCNH2 in patients with long QT syndrome. Heart Rhythm. 2008;5:1275–1281. doi: 10.1016/j.hrthm.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 50.Tester DJ, Benton AJ, Train L, et al. Prevalence and spectrum of large deletions or duplications in the major long QT syndrome-susceptibility genes and implications for long QT syndrome genetic testing. Am J Cardiol. 2010;106:1124–1128. doi: 10.1016/j.amjcard.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kapa S, Tester DJ, Salisbury BA, et al. Genetic testing for long-QT syndrome: distinguishing pathogenic mutations from benign variants. Circulation. 2009;120:1752–1760. doi: 10.1161/CIRCULATIONAHA.109.863076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Priori SG, Schwartz PJ, Napolitano C, et al. Risk stratification in the long-QT syndrome. N Engl J Med. 2003;348:1866–1874. doi: 10.1056/NEJMoa022147. [DOI] [PubMed] [Google Scholar]
- 53.Moss AJ, Zareba W, Hall WJ, et al. Effectiveness and limitations of beta-blocker therapy in congenital long-QT syndrome. Circulation. 2000;101:616–623. doi: 10.1161/01.cir.101.6.616. [DOI] [PubMed] [Google Scholar]
- 54.Moss AJ, Windle JR, Hall WJ, et al. Safety and efficacy of flecainide in subjects with Long QT-3 syndrome (DeltaKPQ mutation): a randomized, double-blind, placebo-controlled clinical trial. Ann Noninvasive Electrocardiol. 2005;10(4 Suppl):59–66. doi: 10.1111/j.1542-474X.2005.00077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruan Y, Liu N, Bloise R, et al. Gating properties of SCN5A mutations and the response to mexiletine in long-QT syndrome type 3 patients. Circulation. 2007;116:1137–1144. doi: 10.1161/CIRCULATIONAHA.107.707877. [DOI] [PubMed] [Google Scholar]
- 56.Priori SG, Napolitano C, Memmi M, et al. Clinical and molecular characterization of patients with catecholaminergic polymorphic ventricular tachycardia. Circulation. 2002;106:69–74. doi: 10.1161/01.cir.0000020013.73106.d8. [DOI] [PubMed] [Google Scholar]
- 57.Medeiros-Domingo A, Bhuiyan ZA, Tester DJ, et al. The RYR2-encoded ryanodine receptor/calcium release channel in patients diagnosed previously with either catecholaminergic polymorphic ventricular tachycardia or geno-type negative, exercise-induced long QT syndrome: a comprehensive open reading frame mutational analysis. J Am Coll Cardiol. 2009;54:2065–2074. doi: 10.1016/j.jacc.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tester DJ, Ackerman MJ. Genetic testing for potentially lethal, highly treatable inherited cardiomyopathies/channelopathies in clinical practice. Circulation. 2011;123:1021–1037. doi: 10.1161/CIRCULATIONAHA.109.914838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krahn AD, Gollob M, Yee R, et al. Diagnosis of unexplained cardiac arrest: role of adrenaline and procainamide infusion. Circulation. 2005;112:2228–2234. doi: 10.1161/CIRCULATIONAHA.105.552166. [DOI] [PubMed] [Google Scholar]
- 60.Postma AV, Denjoy I, Kamblock J, et al. Catecholaminergic polymorphic ventricular tachycardia: RYR2 mutations, bradycardia, and follow up of the patients. J Med Genet. 2005;42:863–870. doi: 10.1136/jmg.2004.028993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61■.van der Werf C, Nederend I, Hofman N, et al. Familial evaluation in catecholaminergic polymorphic ventricular tachycardia: disease penetrance and expression in cardiac ryanodine receptor mutation-carrying relatives. Circ Arrhythm Electrophysiol. 2012;5:748–756. doi: 10.1161/CIRCEP.112.970517. [This study provides a much needed assessment of the penetrance and expressivity of CPVT-causative RYR2 mutations.] [DOI] [PubMed] [Google Scholar]
- 62.Bai R, Napolitano C, Bloise R, et al. Yield of genetic screening in inherited cardiac channelopathies: how to prioritize access to genetic testing. Circ Arrhythm Electrophysiol. 2009;2:6–15. doi: 10.1161/CIRCEP.108.782888. [DOI] [PubMed] [Google Scholar]
- 63.Antzelevitch C, Brugada P, Borggrefe M, et al. Brugada syndrome: report of the second consensus conference – endorsed by the Heart Rhythm Society and the European Heart Rhythm Association. Circulation. 2005;111:659–670. doi: 10.1161/01.CIR.0000152479.54298.51. [DOI] [PubMed] [Google Scholar]
- 64.Brugada P, Brugada J. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: a distinct clinical and electrocardiographic syndrome. A multicenter report. J Am Coll Cardiol. 1992;20:1391–1396. doi: 10.1016/0735-1097(92)90253-j. [DOI] [PubMed] [Google Scholar]
- 65.Chen PS, Priori SG. The Brugada syndrome. J Am Coll Cardiol. 2008;51:1176–1180. doi: 10.1016/j.jacc.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 66.Shimizu W. Clinical impact of genetic studies in lethal inherited cardiac arrhythmias. Circ J. 2008;72:1926–1936. doi: 10.1253/circj.cj-08-0947. [DOI] [PubMed] [Google Scholar]
- 67.Priori SG, Napolitano C, Gasparini M, et al. Clinical and genetic heterogeneity of right bundle branch block and ST-segment elevation syndrome: a prospective evaluation of 52 families. Circulation. 2000;102:2509–2515. doi: 10.1161/01.cir.102.20.2509. [DOI] [PubMed] [Google Scholar]
- 68.Schulze-Bahr E, Eckardt L, Breithardt G, et al. Sodium channel gene (SCN5A) mutations in 44 index patients with Brugada syndrome: different incidences in familial and sporadic disease. Hum Mutat. 2003;21:651–652. doi: 10.1002/humu.9144. [DOI] [PubMed] [Google Scholar]
- 69.Priori SG, Napolitano C, Gasparini M, et al. Natural history of Brugada syndrome: insights for risk stratification and management. Circulation. 2002;105:1342–1347. doi: 10.1161/hc1102.105288. [DOI] [PubMed] [Google Scholar]
- 70■.Crotti L, Marcou CA, Tester DJ, et al. Spectrum and prevalence of mutations involving BrS1- through BrS12-susceptibility genes in a cohort of unrelated patients referred for Brugada syndrome genetic testing: implications for genetic testing. J Am Coll Cardiol. 2012;60:1410–1418. doi: 10.1016/j.jacc.2012.04.037. [Comprehensive assessment of the yield of BrS1 through BrS12 genetic testing in a large and clinically diverse cohort of BrS cases.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kapplinger JD, Tester DJ, Alders M, et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010;7:33–46. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]