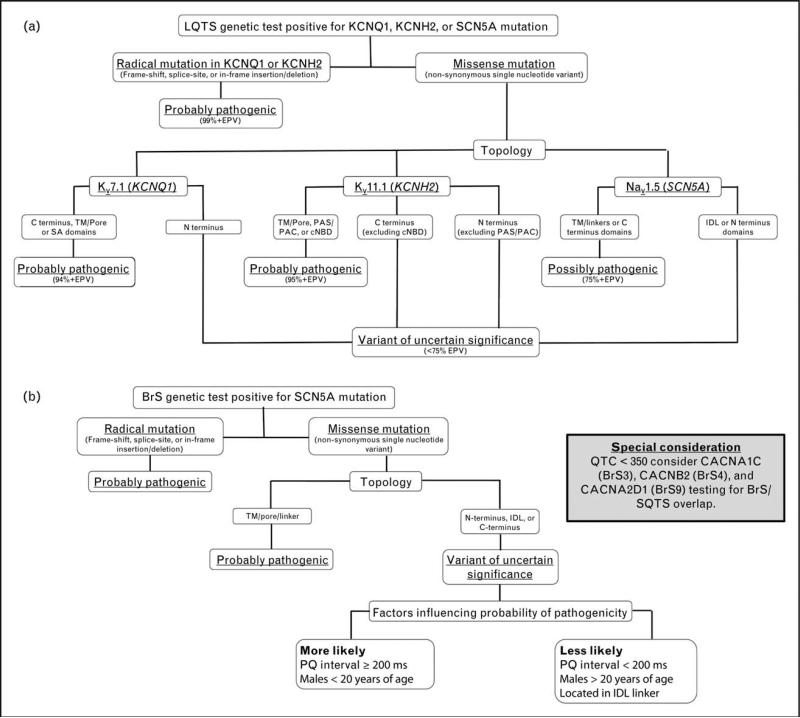
FIGURE 2.
Evidence-based algorithms designed to aid in the interpretation of positive long QT syndrome and Brugada syndrome genetic testing results. (a) Algorithm for interpreting a positive long QT syndrome (LQTS) genetic test. Radical mutations that significantly alter/truncate Kv7.1 or Kv11.1, such as insertions/deletions, alteration of intronic/exonic splice site boundaries, and nonsense mutations, are probably LQTS-associated. Those rare, absent in controls, nonsynonymous single nucleotide variants (nsSNVs; missense mutations) that localize to Kv7.1 (TM/pore, SA, or C terminal domains), Kv11.1 (TM/Pore, PAS/PAC, or cNBD), or Nav1.5 (TM/pore/linker or C terminus) are probably or possibly pathogenic. Variants outside these topological structure-function domains are deemed to be variants of uncertain significance (VUS), unless additional evidence is present (e.g., cosegregation with disease, LQTS-like electrophysiological phenotype, etc.). cNBD, cyclic nucleotide binding domain; EPV, estimated predictive value; IDL, interdomain linker; PAC, per-arnt-sim C-terminal associated; PAS, per-arnt-sim; SA, subunit assembly; TM, transmembrane. (b) Algorithm for interpreting a positive Brugada syndrome (BrS) genetic test. Radical mutations that significantly alter/truncate Nav1.5 protein structure, such as insertions/deletions, alteration of intronic/exonic splice site boundaries, and nonsense mutations, are probably pathogenic. Those rare, absent in controls, nsSNVs (missense mutations) that localize to the Nav1.5 TM/pore/linker are probably pathogenic. Variants outside this topological structure-function domain, particularly those residing in the first interdomain linker (IDL), are of uncertain disease relevance. When such a VUS is identified in an individual with a PQ interval at least 200 ms or a male less than 20 years of age, its probability of pathogenicity increases but not sufficiently so to be declared a BrS1-associated mutation without additional evidence. IDL, interdomain linker; TM, transmembrane domain.