Abstract
We present a PCR-based laboratory exercise that can be used with first- or second-year biology students to help overcome common misconceptions about gene expression. Biology students typically do not have a clear understanding of the difference between genes (DNA) and gene expression (mRNA/protein) and often believe that genes exist in an organism or cell only when they are expressed. This laboratory exercise allows students to carry out a PCR-based experiment designed to challenge their misunderstanding of the difference between genes and gene expression. Students first transform E. coli with an inducible GFP gene containing plasmid and observe induced and un-induced colonies. The following exercise creates cognitive dissonance when actual PCR results contradict their initial (incorrect) predictions of the presence of the GFP gene in transformed cells. Field testing of this laboratory exercise resulted in learning gains on both knowledge and application questions on concepts related to genes and gene expression.
INTRODUCTION
The central dogma of molecular biology was first described by Francis Crick more than 40 years ago when he wrote, “The central dogma of molecular biology deals with the detailed residue-by-residue transfer of sequential information. It states that such information cannot be transferred from protein to either protein or nucleic acid” (8). The canonical interpretation of the Central Dogma is that genetic information (DNA) is transcribed into transient messenger molecules (RNA) that direct synthesis of a particular protein product. Most discussion of the Central Dogma, either by college faculty or textbook authors, includes a drawing, figure or representation that summarizes the Central Dogma similar to the diagram shown in Figure 1.
FIGURE 1.

Typical depiction of the Central Dogma concept.
While this doctrine remained relatively intact for decades, work in the fields of proteomics, genomics, and bioinformatics has produced many exceptions to the rules instated by the Central Dogma. Some DNA codes for functional RNA molecules, not proteins (5,10). Viruses that have an RNA genome must reverse engineer their RNA genome into DNA (3). RNA editing, consisting of structural and coding changes in an RNA molecule, seems to add a detour on the linear path to protein product (19), and prions (infectious proteins) seem to skip most of the path altogether (11). In describing the Encyclopedia of DNA Elements (ENCODE) project, Pennisi (23) eloquently argues that scientists must reconsider the traditional meaning of the term “gene.” When factored in with RNA processing events that occur in eukaryotic cells, such as RNA splicing and alternative splicing, or the feedback loops used to control transcription, it is readily apparent that applying the Central Dogma is anything but simple.
The 2009 Vision and Change report from the American Association for the Advancement of Science and the National Science Foundation identified “Information Flow, Exchange and Storage” as one of the “Core Concepts for Biological Literacy” (2). Topics that fall under this core concept, such as transmission genetics and Central Dogma, typically present problems for college biology students and thus have sparked this call for change in the way that undergraduate biology is taught. The molecular basis of inheritance is a difficult topic for many biology students, as discussed by many researchers in biology education (1, 16–18, 22, 26, 28). One of the most common issues is student misunderstanding about the relationship between genes, alleles and chromosomes. For example, Lewis and Kattman pointed out that many students think of genes as “particles” (15), a phenomenon we have observed in our own work exploring knowledge transfer with advanced biology students (21). Students also mistakenly believe that genes are only present in a cell when they are actively being expressed or are needed for that particular cell (6, 25). For example, they might say that a gene for a neural-specific protein is only present in neurons.
Weak mental models of core concepts related to Central Dogma may lead to intransigent misconceptions about biological processes such as replication, transcription, and translation. More than 25 years ago, Fisher observed that students incorrectly believed protein translation was the process of amino acid synthesis, not the process of building polypeptides from amino acid building blocks (9). More recent work uncovered that biology students who struggle with concepts related to gene structure (e.g. operons) and expression do not connect the significance of the Central Dogma in the context of gene regulation (12).
Through our experience in the classroom and laboratory we have observed confusion between genes (a dedicated stretch of DNA in a given genome or chromosome) and gene expression (transcription and translation of a gene into a functional product) by numerous students in many different contexts. In order to address this confusion, we developed a laboratory exercise that allows students a way to visualize the difference between genes (DNA) and products of expressed genes.
Education research has shown that allowing students to predict results, invent models, or construct a formula before being given the “correct answer” is a powerful way to improve student learning. Schwartz and Bransford, for example, have described increased learning in students who first created graphs to describe data sets from psychology experiments, compared to peers who summarized a chapter on the same experiments (24). One way to increase learning gains, then, is to prime students by allowing them to construct models or predict results before instruction is continued. In a nonmajor biology course, for example, when students were asked to make observations and discuss concepts within their lab groups before presenting their results to the class, students had significantly higher quiz scores, higher attendance rates, and greater appreciation and enjoyment of science than their peers in a traditional lab course (27).
Following this model we have created a laboratory exercise that allows students to predict experimental results that often do not agree with their final data. Learning occurs when students’ misconceptions are challenged by results of a simple, and robust, PCR assay. The technical steps of this laboratory project are not novel; students are performing bacterial transformation, PCR, and gel electrophoresis. We have transformed these standard laboratory procedures, however, into an innovative, constructionist activity that creates cognitive dissonance that requires students to reflect and apply their new knowledge.
Intended audience
Here we present a hands-on laboratory exercise and accompanying assessment strategies that would be appropriate for second-year major-level biology students enrolled in a Cell Biology or Molecular Biology course to increase understanding of the difference between genes (DNA) and gene expression. This exercise may also be appropriate for an Introductory Biology course if it was introduced to the students later in a year-long sequence, once they had had enough time to develop basic molecular techniques. This project could also be adapted for a non-major biology or genetics course, as concepts related to genes and gene expression are crucial for genetics literacy from a health and public policy perspective (13, 14, 20). This laboratory exercise has been written to accommodate students working singly or in teams, and can easily be scaled up to accommodate multiple laboratory sections, which are usually capped at 20–24 students.
Learning time
The project was designed to last for a total of three laboratory periods, where each session takes less than two hours to complete. During the first session, students transform E. coli (HB101-K12, a common laboratory strain which poses minimal risk to students) with pGLO (a plasmid containing GFP under control of the araBAD promoter) and plate on selective media (Luria Broth agar containing ampicillin, or “LB amp”) as well as a negative control without antibiotic (LB). They also complete a tutorial on PCR methodology (Appendix 1) to prepare them for the second session. Before the second session, they observe growth of colonies from their transformation plates and streak them on LB amp plates with and without arabinose in the media. In the second session they observe the plates under ultraviolet light to see green fluorescent protein (GFP) expression, and perform PCR (polymerase chain reaction) directly on the colonies (Appendix 2). They also must complete a short free-response preassessment quiz (graded for effort, not correctness) before they leave the laboratory for the day in order to elicit reflection about the experiment (Appendix 3). In the third session, students run their products on an agarose gel and analyze their results. Finally, they complete a short postlaboratory reflection (Appendix 4) in order to confront their misconceptions and promote synthesis of the information. It is important that the instructor not “give away” the answer to students during pre-lab discussion. Students need to grapple with the concepts on their own in order to construct new knowledge.
Prerequisite student knowledge
This laboratory exercise is intended to be used with students who have introductory knowledge of the Central Dogma (DNA replication, RNA transcription, and protein translation), which is why this exercise may not be appropriate for a nonmajor biology course. Familiarity with bacterial operon systems would be helpful, but this is not an absolute requirement as long as some time is spent discussing essential components of transcriptional control as a type of pre-lab discussion. Familiarity with basic microbiology techniques (streaking and spreading) is helpful but not necessary, as long as the instructor demonstrates and discusses these techniques. Basic knowledge about PCR and its utility in molecular biology research is also helpful but not strictly necessary. A brief pre-lab introduction by the instructor may suffice, since the first session includes an exercise that walks the students through the process and what happens at a molecular level at each step. The materials do assume, however, that students have already learned about the mechanism of in vivo DNA replication, and therefore have an understanding of the components required in a cell and their basic functions.
Students should be able to use a micropipettor correctly, solve simple dilution problems, and work semi-independently in the lab (i.e. be able to carry out a protocol and use safe laboratory practices). It is also recommended that students already be familiar with electrophoresis of DNA on agarose gels. If not, instructors may need to spend some time on the concept, perhaps incorporating other materials about the method and practice loading gels.
Learning objectives
At the completion of this laboratory exercise students should be able to:
Apply knowledge that information in DNA is permanent and information in mRNA is transient.
Predict results from a PCR experiment in which a gene is present in a cell but not expressed.
Explain how the results of gene expression can be observed as a phenotypic change.
PROCEDURE
The laboratory exercise as written requires three laboratory sessions, plus a little work in between labs. It would also be possible to skip the first session, instead starting with strains that already contain pGLO streaked on LB amp and LB amp ara (LB with both ampicillin and arabinose) plates.
Part I: Transformation of E. coli with inducible pGLO plasmid (based on Bio-Rad or other transformation protocol). Completion of PCR tutorial (Appendix 1).
Part II: Comparison of GFP expression in arabinose-induced and uninduced cultures. Direct PCR on colonies to amplify GFP gene fragment and prediction of PCR results (Appendices 2 and 3).
Part III: Agarose gel electrophoresis of PCR products and completion of postlab reflective assignment (Appendix 4).
The pGLO plasmid and E. coli HB101-K12 can be purchased separately or as part of the pGLO Bacterial Transformation Kit (Bio-Rad). The pGLO Bacterial Transformation Kit has been designed for use in high schools or in settings that are resource-limited; instructors may wish to purchase only the plasmid and host E. coli strain and to prepare the 50 mM CaCl2 solution, LB media +/− ampicillin (100 μg/ml) and +/− L-arabinose (0.3% w/v) broths and plates themselves.
There are minimal safety issues associated with this exercise. Students should practice safe laboratory behavior and wear laboratory coats and disposable gloves when handling the E. coli.
Student instructions
Students should transform E. coli with the inducible pGLO plasmid (instructions available at Bio-Rad.com), and plate aliquots on LB, LB amp and LB amp ara. After incubation at 37°C for one day, they should compare the number of colonies observed on each plate and examine them under an ultraviolet light source to confirm presence of GFP. If desired, students can streak single colonies on new plates before the next lab session; we recommend using the same colony for streaking on LB amp and LB amp ara to drive home the idea that the same cells express different genes on different media. In the next scheduled lab session students write and implement a PCR protocol to try and amplify the GFP gene directly from bacteria grown on the three different plates (LB, LB amp, LB amp ara).
Students work independently, or in pairs, to create a suitable PCR protocol using guidelines supplied by the instructor (Appendix 2). Students must present a detailed PCR protocol to a teaching assistant or instructor for approval before setting up the actual PCR. The instructor should hold all PCR tubes on ice until all students have completed the setup, and run all samples together in a thermocycler. Note that extended time on ice may result in strong primer-dimer bands in the final product.
Students are given a worksheet to complete (independently) after they have finished setting up their PCR. Students are reminded that worksheets are not graded for correctness, just effort (a small number of points are given to ensure compliance), and will be useful for them to use for comparison when they visualize their actual PCR results using agarose gel electrophoresis.
In the final session, students run 10 μl of each of their PCR products on a 1% agarose gel.
Instructor notes
Several suggestions will ensure this laboratory exercise runs as intended.
Students generally respond much more enthusiastically and write more when given an assessment that is graded for effort and not correctness. When students are allowed to fully explain their thoughts, without fear of losing points for incorrect thinking, it becomes easier for the instructor to understand students’ mental models of replication and expression. It is also crucial that students be asked to explain their PCR predictions with words, and not just have the students draw bands on a blank gel. When students have very little experience with PCR they may be unsure as to what results would actually look like after gel electrophoresis. If only drawings are solicited it may be difficult for an instructor to gauge whether the students are confused about replication vs. transcription or if they are just not experienced enough to accurately represent PCR bands on a representation of an agarose gel.
Lastly, it is crucial that instructors ask each student (or pair of students) to analyze and explain their PCR results out loud once they see their final gels, as this provides an opportunity for students to confront their misunderstanding of or misconceptions about replication of genes versus transcription of genes. This pedagogical tactic may help to break down flawed mental models of these two processes as described in Science Teaching Reconsidered (7). If students are allowed to leave the lab without having to explain their results, they might assume their experiment did not work correctly or they may leave without thinking about what actually happened.
Suggestions
Although PCR is typically an easy procedure for experts and experienced research students, we do not assume the same to be true when working with inexperienced undergraduate students. Instructors should stress proper techniques to prevent contamination between samples (changing pipette tips often, using filter tips, keeping tube lids closed between transfers, etc.).
Very short non-specific PCR artifacts (“primer-dimers”) may be observable after agarose electrophoresis in all lanes containing PCR products. Since there is a large difference in size between the primer-dimers (20–40 bp) and the actual PCR product (714 bp) these artifacts present students with another chance to critically analyze their results. We suggest that instructors ask students to determine the size of both the primer-dimers and the PCR products while visualizing the DNA gels to help them realize the large difference in size. Instructors may also ask students to come up with a logical explanation for the appearance of very small products that show up in all experimental lanes. Lastly, PCR artifacts are commonly observed using traditional methods, so there is benefit to providing real-life data to undergraduate students. Depending on time and availability of reagents, the instructor might even ask for volunteers to optimize PCR conditions to decrease primer-dimer artifacts (e.g. using less primer in PCR) in future lab sessions.
DISCUSSION
This laboratory exercise was carried out with a second-year Molecular Biology class (n = 49) comprised of multiple laboratory sections. In Part I, students transformed E. coli with the inducible pGLO plasmid (using a modified protocol from Bio-Rad) and plated transformants on LB amp +/− arabinose and observed GFP production from induced pGLO E. coli. In Part II, students carried out PCR assays and completed the first pre-lab assessment (Appendix 3), and in Part III, students analyzed their PCR products using DNA gel electrophoresis. Postlaboratory assessments, also described in Appendix 3, were used to assess student learning.
Field testing
All student data presented here were gathered following institutional review board guidelines. Before seeing the results of their PCR, students in the winter section (n = 49) were given an open-ended assessment (Appendix 3, Pre-lab assessment) used to measure baseline knowledge of Learning Objective 1 (Apply knowledge that information in DNA is permanent and information in mRNA is transient) and Learning Objective 2 (Predict results from a PCR experiment in which a gene is present in a cell but not expressed). Students were asked to draw and explain their predicted PCR results in an open-ended assessment which was only graded on effort.
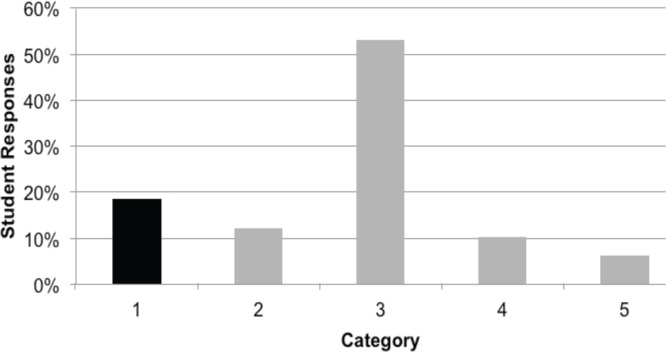
Student written responses were coded for correctness with respect to demonstrating knowledge that DNA is permanent (e.g. both transformed strains contain the gene for GFP; Learning Objective 1) and gel drawing for prediction of PCR results that reflected this fact (Learning Objective 2). The results of this preassessment confirmed that most students do not have a solid understanding of either PCR (replication) or gene expression (Table 1). Figure 2 presents the breakdown of student responses, showing only a fraction of students could clearly describe that a gene is present regardless of its expression.
TABLE 1.
Correlation of pre- and postlaboratory assessments with learning objectives and learning gains.
| Assessment |
Learning Objective
|
Learning Gain | ||
|---|---|---|---|---|
| Apply knowledge that information in DNA is permanent and information in mRNA is transient | Predict results from a PCR experiment in which a gene is present in an organism but not expressed | Explain how the results of gene expression can be observed as a phenotypic change | ||
| Pre-lab: Students were asked to draw and explain their predicted PCR results in open-ended format (Appendix 3, Pre-lab assessment) | 18% of students answered correctly | 18% of students answered correctly | ----- | ----- |
| Postlab: Students were asked apply knowledge that DNA is permanent and mRNA is transient by recognizing experimental results of a PCR assay (MCQ format) (Appendix 3, Postlab assessment 1) | 56% of students answered correctly | ------ | ------ | 46.3% |
| Postlab: Students were asked to predict the results of a PCR experiment (MCQ format) using DNA from induced and uninduced experimental organisms (Appendix 3, Postlab assessment 2) | ------ | 52% of students answered correctly | ----- | 41.5% |
| Postlab: Students examined an image of pGLO-transformed E. coli +/− arabinose and were asked to explain differences and similarities at the level of DNA and protein between the two strains (Appendix 3, Postlab assessment 3) | ------ | ------ | 85% of students answered correctly | ---- |
FIGURE 2.
Sophomore-level biology students do not demonstrate a clear understanding of PCR or the difference between DNA replication and expression. Students from a second-year Molecular Biology course (n = 49) predicted the results of their PCR experiment with the open-ended question shown in Appendix 3 (Pre-lab assessment). Categories of student responses are as follows: 1) Gene (DNA) is present whether or not it is being expressed; 2) Amount of gene (DNA) present is proportional to expression level of gene; 3) Gene (DNA) is only present when it is being expressed; 4) Presence of arabinose allows the GFP gene to get added to the E. coli genome; 5) Unable to follow logic.
Only 18% (9/49) of the students provided a correct argument for why a fragment of the GFP gene would be amplified in pGLO-transformed E. coli independent of growth conditions. Students in this group provided reasoning such as:
“It doesn’t matter where the E. coli are grown; if they have the pGLO plasmid they have the GFP gene, and it will be amplified.”
“Wild type has no plasmid for GFP . . . the E. coli without arabinose would still have the plasmid even if unexpressed.”
Six of the 49 students (12%) explained that more GFP gene would be amplified in the arabinose-induced pGLO E. coli than in the uninduced and drew one faint band and one heavy band on their gel representations. These students described a scenario in which gene expression was proportional to the amount of the particular gene and provided these explanations for why less PCR product would be visible using template from uninduced pGLO E. coli DNA:
“The sans arabinose sample will sporadically express the product.”
“There is not much pGLO due to the lack of arabinose.”
“[N]ot all [of the GFP] would be expressed.”
Twenty-six (53%) of the students stated that a fragment of the GFP gene would only be amplified from induced pGLO E. coli DNA. Student explanations revealed that this population thought that the GFP gene was only present when it was being expressed.
“Without arabinose the enzyme is not activated.”
“Only GFP is expressed with [when] arabinose is present.”
“GFP gene cannot be targeted under these conditions.”
“The E. coli grown without arabinose will not have the promoter region exposed and will not be duplicated.”
Five additional students also thought that the GFP gene fragment would only be amplified from induced pGLO E. coli DNA but indicated that the PCR product would somehow get added to the E. coli genome (extending the size of the genome). These students were most likely struggling with the idea of PCR, and what results on an agarose gel would look like, and offered explanations such as:
“The pGLO + arabinose will have more base-pairs than that of pGLO – arabinose.”
“The genome of pGLO + arabinose will get progressively larger [compared to wild-type and DNA from uninduced transformed E. coli].”
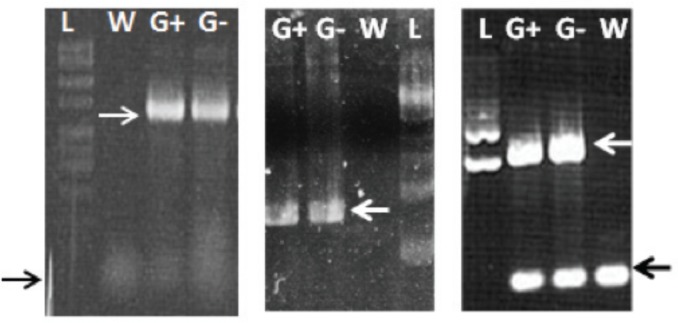
When students returned to the lab the following week they ran 8 μl of their PCR products in 1.0% agarose gels (Fig. 3). Primer-dimers are often seen but the large size difference between true product and artifact allows students to differentiate between the two.
FIGURE 3.
PCR amplification of the GFP gene from pGLO E. coli. Examples of three student gels demonstrating GFP gene amplification (white arrows at 714 bp) in samples labeled G+ (DNA from induced pGLO E. coli) and G– (DNA from uninduced pGLO E. coli) but not W (DNA from wild-type E. coli). DNA ladders (L) are included on all gels. White arrows indicate the 714 bp PCR product. Black arrows indicate primer-dimer product, which varies in intensity depending on conditions.
While students are photographing or analyzing their gels it is imperative that the lab instructor ask them to explain their results (see Instructor notes section). Students who correctly predicted the PCR results (about 18% of our population) typically are not surprised by their own results but are generally pleased, nonetheless.
Students who think they will amplify the fragment of the GFP gene only from induced pGLO E. coli, or think they will see a large difference in the amount of DNA amplified from the induced vs. uninduced sample DNA, are usually perplexed when they see their PCR gels. Many students, in fact, say that they must have done something incorrectly because they should only see one band, not two, or one heavy band and one light band. The instructor can address this challenge to students’ mental models by either:
Sending the students to talk with other students in the lab to compare their results before returning to the instructor with a new explanation.
Probing the students to explain why they thought there would be a difference in outcome when induced or uninduced sample DNA was used as template.
Asking the students why the presence of arabinose would have an effect on their assay that involves replication.
Once students realize that the gene for GFP (the DNA) is represented in both induced and uninduced transformed E. coli DNA the instructor can point out the difference in their thinking before and after they were able to visualize their PCR results. Many students verbally acknowledge they “got this question wrong” on the previous quiz and admit the laboratory exercise was actually useful.
After visualizing the results of the PCR assay, students complete a short reflective assignment where they compare and contrast their predicted results with their actual results (Appendix 4).
Evidence of student learning
Different assessment strategies were used to measure student learning (Table 1), and postlaboratory gains were calculated using the formula: (Postlab assessment score – Pre-lab assessment score)/(100% – Pre-lab assessment score). Students were presented with two multiple-choice questions (MCQs): one on an in-class exam approximately one week after the laboratory was completed and the other on the final examination that occurred two weeks after the laboratory project. To measure learning gains from Learning Objective 1, students were presented with an exam question (MCQ format) in which they had to apply knowledge that DNA is permanent and expression is transient by recognizing experimental results of a PCR assay (Appendix 3, Postlab assessment 1). Fifty-six percent of the students who had completed the laboratory exercise could correctly answer this question, resulting in a 46% learning gain (Table 1). To measure learning gains for Learning Objective 2, students were asked to predict the results of a PCR experiment (MCQ format) using DNA from induced and uninduced experimental organisms (Appendix 3, Postlab assessment 2). Fifty-two percent of the students who had completed the laboratory exercise could correctly answer this question, resulting in a 41.5% learning gain (Table 1).
On a laboratory practical examination students were shown an image of pGLO-transformed E. coli growing in the presence or absence of L-arabinose and asked whether both strains contained the GFP gene (Appendix 3, Postlab assessment 3). This question tests whether students are able to recognize that the results of gene expression can be observed as a phenotypic change (Learning Objective 3). Approximately 85% of tested students (Table 1) answered correctly, using explanations such as:
“It just might not be actively expressed if no arabinose is present.”
“They both contain the gene that codes for GFP, but only GFP is expressed in the presence of arabinose.”
“The genetic instruction is there on both but is not being expressed on the plate that is missing arabinose.”
Although we did not have a preassessment for Learning Objective 3 (Explain how the results of gene expression can be observed as a phenotypic change), the underlying concept is related to the previous learning objectives and it can be reasonably inferred that students have minimal prior understanding of the concept. We cannot calculate learning gains for this particular objective, but the students did demonstrate high performance on the open-ended assessment question. Additionally, only 44% of incoming Molecular Biology students correctly answered that various human cell types would all contain the gene for a liver-specific enzyme. The core idea of this question relates to Learning Objective 3; in other words, a gene may only be expressed under certain conditions to produce a specific phenotype (e.g. liver-enzymatic activity). Thus, the fact that 85% of tested students were able to answer the posttest question correctly appears to be a significant learning gain.
Overall, our test population demonstrated pronounced learning gains using this laboratory exercise that allows students to generate and analyze data that challenges their initial ideas about genes and gene expression. We have transformed several standard molecular biology exercises into an innovative, constructionist activity that can be used with a variety of student populations. As reviewed by Baviskar (4), a constructivist activity: 1) elicits prior knowledge, 2) creates cognitive dissonance, 3) applies new knowledge with feedback, and 4) allows for student reflection on learning. Table 2 provides a summary of how this laboratory exercise meets the criteria. The students also seemed to enjoy the laboratory experiment and felt that it was useful in helping them learn. Students wrote that:
“. . . the DNA from induced and uninduced both appeared on the gel at the same distance. I think it’s a good way to illustrate that when a protein is overexpressed, the quantity of the DNA is not affected.”
“It did give me a visual of what we were trying to understand.”
“When I saw the gel it all came together for me. I got the difference between genes and gene expression.”
TABLE 2.
Comparison of the laboratory exercise to features of a constructivist activity (as described by Baviskar (4)).
| Steps in Constructivist Activity | In Context of Laboratory Exercise |
|---|---|
| Eliciting prior knowledge | Open-ended assessment after completion of Part II (see Appendix 3) |
| Creating cognitive dissonance | Visualization of PCR results that are not predicted by ∼80% of student population |
| Applying new knowledge with feedback | Discussion with laboratory instructor about PCR results |
| Allowing student reflection on learning | Reflection questions after completion of Part III (see Appendix 4) |
Possible modifications
Although this laboratory exercise has been optimized to be carried out in the context of arabinose-induced bacterial GFP expression, any inducible system that the instructor is familiar with would presumably be suitable. GFP-positive induced E. coli cultures offer the distinct advantage of a fluorescent protein product that can easily be visualized using UV illumination. This exercise can easily be scaled up to accommodate a number of laboratory sections within the same course. Reagents and their amounts required are listed in Appendix 2. Although primer sequences are provided, an instructor could expand the exercise and include a primer-design component.
Another suggestion is to have the students isolate RNA from cells and perform a semi-quantitative RT-PCR assay to demonstrate differences in mRNA levels. Alternatively, protein could be isolated and used for Western blot analysis with anti-GFP antibodies. Students could also visualize GFP using protein purification columns (Bio-Rad) to clarify the difference between presence of a gene and presence of its protein product.
SUPPLEMENTAL MATERIALS
Appendix 1: Student lab protocol
Appendix 2: Instructor notes
Appendix 3: Pre- and postlab assessment questions
Appendix 4: Reflection activity
Acknowledgments
The authors declare that there are no conflicts of interest.
REFERENCES
- 1.Allchin D. Mending mendelism. Am Biol Teach. 2000;62:633–639. doi: 10.1662/0002-7685(2000)062[0632:MM]2.0.CO;2. [DOI] [Google Scholar]
- 2.American Association for the Advancement of Science . Vision and change in undergraduate biology education: a call to action. American Association for the Advancement of Science; Washington, DC: 2009. [Google Scholar]
- 3.Baltimore D. Viruses, viruses, viruses. Eng Sci. 2004;67:20–29. [Google Scholar]
- 4.Baviskar SN, Hartle RT, Whitney T. Essential criteria to characterize constructivist teaching: derived from a review of the literature and applied to five constructivist-teaching method articles. Int J Sci Educ. 2009;31:541–550. doi: 10.1080/09500690701731121. [DOI] [Google Scholar]
- 5.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowling BV, et al. Development and evaluation of a genetics literacy assessment instrument for undergraduates. Genetics. 2008;178:15–22. doi: 10.1534/genetics.107.079533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Committee on Undergraduate Science Education . Science teaching reconsidered: a handbook. The National Academy Press; Washington, DC: 1997. [Google Scholar]
- 8.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 9.Fisher K. A misconception in biology: Amino acids and translation. J Res Sci Teach. 1985;22:3–62. doi: 10.1002/tea.3660220105. [DOI] [Google Scholar]
- 10.Gerstein MB, et al. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- 11.Hunter N. Prion diseases and the central dogma of molecular biology. Trends Microbiol. 1999;7:265–266. doi: 10.1016/S0966-842X(99)01543-7. [DOI] [PubMed] [Google Scholar]
- 12.Khodor J, Halme DG, Walker GC. A hierarchical biology concept framework: a tool for course design. Cell Biol Educ. 2004;3:111–121. doi: 10.1187/cbe.03-10-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolstø SD. Scientific literacy for citizenship: tools for dealing with the science dimension of controversial socioscientific issues. Sci Educ. 2001;85:291–310. doi: 10.1002/sce.1011. [DOI] [Google Scholar]
- 14.Lanie AD, et al. Exploring the public understanding of basic genetic concepts. J Gen Couns. 2004;13:305–320. doi: 10.1023/B:JOGC.0000035524.66944.6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis J, Kattmann U. Traits, genes, particles and information: re–visiting students’ understandings of genetics. Int J Sci Educ. 2004;26:195–206. doi: 10.1080/0950069032000072782. [DOI] [Google Scholar]
- 16.Lewis J, Leach J, Wood-Robinson C. All in the genes?—young people’s understanding of the nature of genes. J Biol Educ. 2000;34:74–79. doi: 10.1080/00219266.2000.9655689. [DOI] [Google Scholar]
- 17.Lewis J, Wood-Robinson C. Genes, chromosomes, cell division and inheritance—do students see any relationship? Int J Sci Educ. 2000;22:177–195. doi: 10.1080/095006900289949. [DOI] [Google Scholar]
- 18.Marbach-Ad G. Attempting to break the code in student comprehension of genetic concepts. J Biol Educ. 2001;35:183–189. doi: 10.1080/00219266.2001.9655775. [DOI] [Google Scholar]
- 19.Maydanovych O, Beal P. Breaking the central dogma by RNA editing. Chem Rev. 2006;106:3397–3411. doi: 10.1021/cr050314a. [DOI] [PubMed] [Google Scholar]
- 20.Miller JD. The measurement of civic scientific literacy. Public Und Sci. 1998;7:203–223. doi: 10.1088/0963-6625/7/3/001. [DOI] [Google Scholar]
- 21.Newman DL, Catavero C, Wright LK. Students fail to transfer knowledge of chromosome structure to topics pertaining to cell division. CBE Life Sci Educ. 2012;11:425–436. doi: 10.1187/cbe.12-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pashley M. A-level students: their problems with gene and allele. J Biol Educ. 1985;28:120–127. doi: 10.1080/00219266.1994.9655377. [DOI] [Google Scholar]
- 23.Pennisi E. DNA study forces rethink of what it means to be a gene. Science. 2007;316:1556–1557. doi: 10.1126/science.316.5831.1556. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz DL, Bransford JD. A time for telling. Cogn Instr. 1998;16:475–522. doi: 10.1207/s1532690xci1604_4. [DOI] [Google Scholar]
- 25.Smith MK, Wood WB, Knight JK. The genetics concept assessment: a new concept inventory for gauging student understanding of genetics. CBE Life Sci Educ. 2008;7:422–430. doi: 10.1187/cbe.08-08-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart J, Hafner B, Dale M. Students’ alternate views of meiosis. Am Biol Teach. 1990;52:228–232. doi: 10.2307/4449090. [DOI] [Google Scholar]
- 27.Travis H, Lord T. Traditional and constructivist teaching techniques: comparing two groups of undergraduate nonscience majors in a biology lab. J Coll Sci Teach. 2004;34:12–18. [Google Scholar]
- 28.Wood-Robinson C, Lewis J, Leach J. Young people’s understanding of the nature of genetic information in the cells of an organism. J Biol Educ. 2000;35:29–36. doi: 10.1080/00219266.2000.9655732. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 1: Student lab protocol
Appendix 2: Instructor notes
Appendix 3: Pre- and postlab assessment questions
Appendix 4: Reflection activity