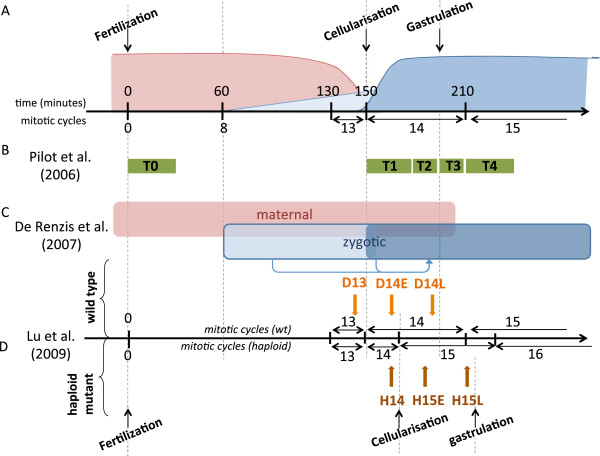
Figure 1.
Temporal organization of Zygotic Genome Activation and design of published high-throughput experiments used to extract early induced gene clusters.(A) Schematic representation of mRNA concentration evolution during early drosophila embryogenesis. The horizontal axis represents time, in minutes after fertilisation (upper scale) or mitotic cycles (lower scale). Red: maternal mRNAs; light blue: first wave of zygotic transcription; dark blue: second wave of zygotic transcription. (B) Time points sampled in transcriptome microarray experiments performed by Pilot et al. (2006). (C) Respective contribution of maternal and zygotic mRNAs. Arrows represent the action of early expressed TF on secondary targets. (D) Time points of transcriptome microarray experiments published by Lu et al. (2009). D and H denote diploids and haploids; the numbers indicate the mitotic cycle number; E and L stand for early and late. Developmental milestones are indicated for haploid mutant embryos; notice the differences with wild-type timing in (A).