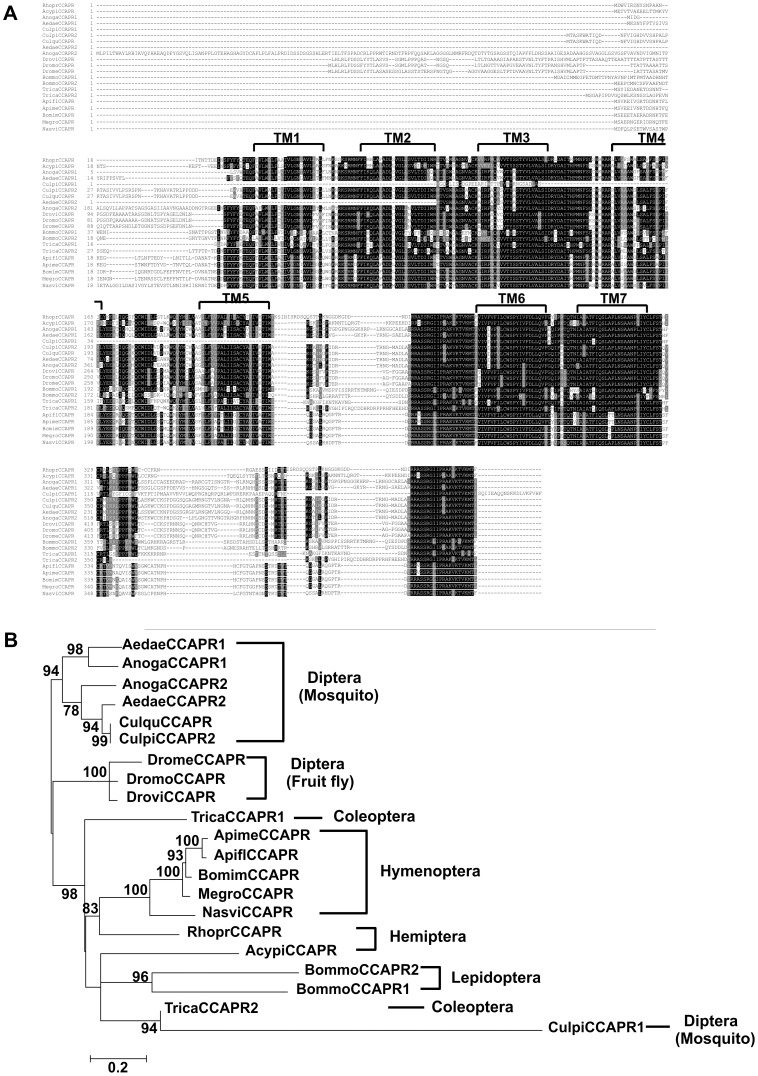
Figure 2. Protein alignment and phylogenetic analysis of CCAP receptors in insects.
A) Amino acid sequence alignment of the CCAP receptors identified or predicted in 21 species in arthropods. The predicted location of the seven transmembrane regions (TM1-TM7) are indicated above each row. Following the 50% majority rule, identical amino acids are shaded in black, and similar amino acids are shaded in gray in column consensus residues. (B) The phylogenetic relationship of the insect CCAP receptors was generated using the Maximum Likelihood method based on the Jones et al. (1992) with frequency model [45]. The numbers at the nodes represent percentage support in 1500 bootstrap replicates. All positions containing gaps and missing data were eliminated. This phylogenetic tree is drawn to scale and the branch lengths are measured in the number of substitutions per site.