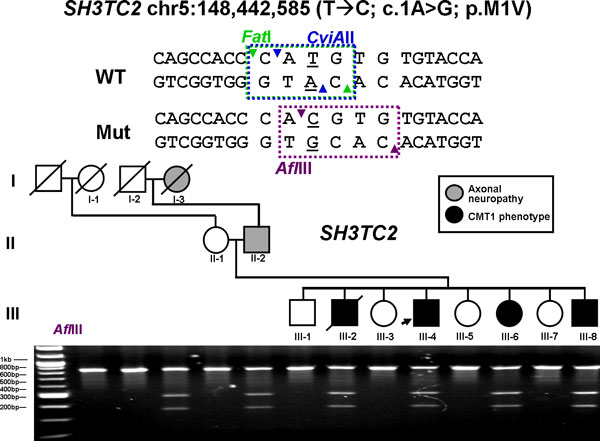
Figure 7.
Flanking sequence of the genomic mutation T → C at position chr5:148,422,778 in SH3TC2. The coding SNV transition c.1A>G causes a substitution of valine for methionine at the initiation codon. An alternative in frame ATG codon is present 90 codons downstream with a less conserved Kozak sequence than the upstream initiation codon. The wild-type allele can be recognized by two different restriction enzymes, FatI and CviAII, but the mutation destroys the restriction site (data not shown). Conversely, transition T→C creates a restriction site that can be recognized by the endonuclease AflIII. The mutation segregates with the axonal neuropathy phenotype and co-segregates as a complex allele (p.M1?; Y169H) with the nonsense mutation p.R954X to cause the CMT phenotype in this family.