Table 1.
«Conserved» miRNA families in the three model legumes.
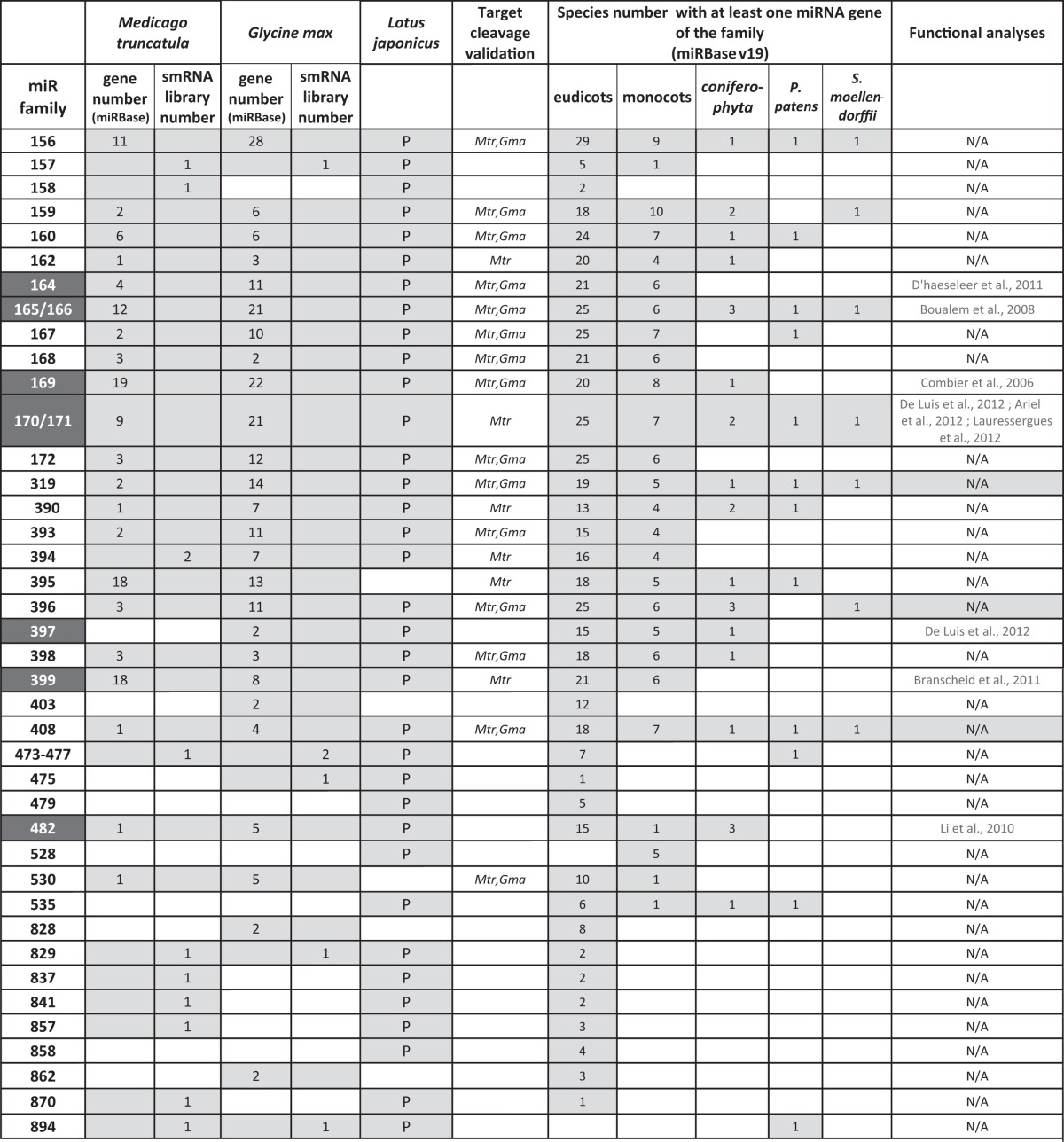
For each conserved miRNA family, the number of genes listed in miRBAse v19 in Medicago truncatula and soybean (Glycine max) are given. If one miRNA was absent from miRBAse, we searched it in smRNA libraries or genome reports and indicated the number of reports where it was identified. References of the smRNA sequencing and genome reports used for these additional searches are (Jagadeeswaran et al., 2009; Kulcheski et al., 2011; Lelandais-Briere et al., 2009; Li et al., 2011; Radwan et al., 2011; Szittya et al., 2008; Turner et al., 2012; Wong et al., 2011; Young et al., 2011; Zhai et al., 2011). Presence in L. japonicus according De Luis et al. (2012) is indicated by P. Validation of target cleavage by degradome experiments in M. truncatula and/or G. max were extracted from the following reports: (Devers et al., 2011; Kulcheski et al., 2011; Song et al., 2012; Turner et al., 2012; Zhai et al., 2011; Zhou et al., 2012); N/A, not applicable. P. patens: Physcomitrella patens (moss); S. moellendorfii: Selaginella moellendorfii (lycophyta) Pale grey boxes: presence of the miRNA, dark gray boxes: miRNA with functional analyses in legumes.
